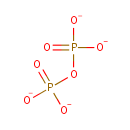
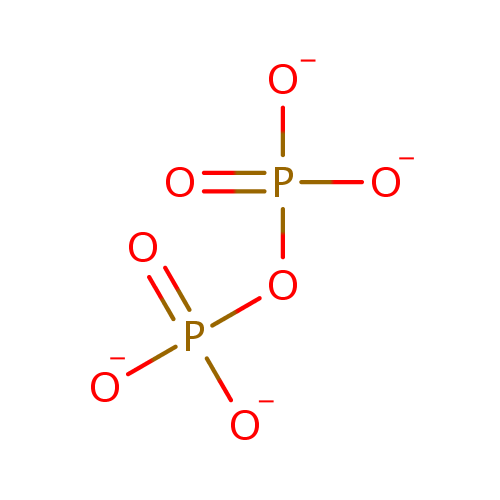
Pyrophosphate (PAMDB000633)
Enzymes
- General function:
- Involved in nucleic acid binding
- Specific function:
- In addition to polymerase activity, this DNA polymerase exhibits 3' to 5' and 5' to 3' exonuclease activity. It is able to utilize nicked circular duplex DNA as a template and can unwind the parental DNA strand from its template
- Gene Name:
- polA
- Locus Tag:
- PA5493
- Molecular weight:
- 99.8 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in anthranilate phosphoribosyltransferase activity
- Specific function:
- Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate
- Gene Name:
- trpD
- Locus Tag:
- PA0650
- Molecular weight:
- 37.4 kDa
Reactions
| Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate. |
| N-(5-phospho-D-ribosyl)-anthranilate + diphosphate = anthranilate + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in adenylate cyclase activity
- Specific function:
- ATP = 3',5'-cyclic AMP + diphosphate
- Gene Name:
- cyaA
- Locus Tag:
- PA5272
- Molecular weight:
- 108.7 kDa
Reactions
| ATP = 3',5'-cyclic AMP + diphosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-tryptophan + tRNA(Trp) = AMP + diphosphate + L-tryptophyl-tRNA(Trp)
- Gene Name:
- trpS
- Locus Tag:
- PA4439
- Molecular weight:
- 49 kDa
Reactions
| ATP + L-tryptophan + tRNA(Trp) = AMP + diphosphate + L-tryptophyl-tRNA(Trp). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of isoleucine to tRNA(Ile). As IleRS can inadvertently accommodate and process structurally similar amino acids such as valine, to avoid such errors it has two additional distinct tRNA(Ile)-dependent editing activities. One activity is designated as 'pretransfer' editing and involves the hydrolysis of activated Val-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Val-tRNA(Ile)
- Gene Name:
- ileS
- Locus Tag:
- PA4560
- Molecular weight:
- 105.5 kDa
Reactions
| ATP + L-isoleucine + tRNA(Ile) = AMP + diphosphate + L-isoleucyl-tRNA(Ile). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Edits incorrectly charged Ser-tRNA(Ala) and Gly- tRNA(Ala) but not incorrectly charged Ser-tRNA(Thr)
- Gene Name:
- alaS
- Locus Tag:
- PA0903
- Molecular weight:
- 94.7 kDa
Reactions
| ATP + L-alanine + tRNA(Ala) = AMP + diphosphate + L-alanyl-tRNA(Ala). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Is required not only for elongation of protein synthesis but also for the initiation of all mRNA translation through initiator tRNA(fMet) aminoacylation
- Gene Name:
- metG
- Locus Tag:
- PA3482
- Molecular weight:
- 74.9 kDa
Reactions
| ATP + L-methionine + tRNA(Met) = AMP + diphosphate + L-methionyl-tRNA(Met). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly)
- Gene Name:
- glyQ
- Locus Tag:
- PA0009
- Molecular weight:
- 36.1 kDa
Reactions
| ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly). |
- General function:
- Involved in arginine-tRNA ligase activity
- Specific function:
- ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly)
- Gene Name:
- glyS
- Locus Tag:
- PA0008
- Molecular weight:
- 74 kDa
Reactions
| ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-glutamine + tRNA(Gln) = AMP + diphosphate + L-glutaminyl-tRNA(Gln)
- Gene Name:
- glnS
- Locus Tag:
- PA1794
- Molecular weight:
- 62.9 kDa
Reactions
| ATP + L-glutamine + tRNA(Gln) = AMP + diphosphate + L-glutaminyl-tRNA(Gln). |
- General function:
- Involved in DNA binding
- Specific function:
- DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. The epsilon subunit contain the editing function and is a proofreading 3'-5' exonuclease
- Gene Name:
- dnaQ
- Locus Tag:
- PA1816
- Molecular weight:
- 26.8 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in GMP synthase (glutamine-hydrolyzing) activity
- Specific function:
- Catalyzes the synthesis of GMP from XMP
- Gene Name:
- guaA
- Locus Tag:
- PA3769
- Molecular weight:
- 58 kDa
Reactions
| ATP + xanthosine 5'-phosphate + L-glutamine + H(2)O = AMP + diphosphate + GMP + L-glutamate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of glutamate to tRNA(Glu) in a two-step reaction:glutamate is first activated by ATP to form Glu-AMP and then transferred to the acceptor end of tRNA(Glu)
- Gene Name:
- gltX
- Locus Tag:
- PA3134
- Molecular weight:
- 56.7 kDa
Reactions
| ATP + L-glutamate + tRNA(Glu) = AMP + diphosphate + L-glutamyl-tRNA(Glu). |
- General function:
- Involved in lipopolysaccharide biosynthetic process
- Specific function:
- Activates KDO (a required 8-carbon sugar) for incorporation into bacterial lipopolysaccharide in Gram-negative bacteria
- Gene Name:
- kdsB
- Locus Tag:
- PA2979
- Molecular weight:
- 27.6 kDa
Reactions
| CTP + 3-deoxy-D-manno-octulosonate = diphosphate + CMP-3-deoxy-D-manno-octulosonate. |
- General function:
- Involved in biotin-[acetyl-CoA-carboxylase] ligase activity
- Specific function:
- BirA acts both as a biotin-operon repressor and as the enzyme that synthesizes the corepressor, acetyl-CoA:carbon-dioxide ligase. This protein also activates biotin to form biotinyl-5'- adenylate and transfers the biotin moiety to biotin-accepting proteins
- Gene Name:
- birA
- Locus Tag:
- PA4280
- Molecular weight:
- 34 kDa
Reactions
| ATP + biotin + apo-[acetyl-CoA:carbon-dioxide ligase (ADP-forming)] = AMP + diphosphate + [acetyl-CoA:carbon-dioxide ligase (ADP-forming)]. |
- General function:
- Involved in nucleotide binding
- Specific function:
- The gamma chain seems to interact with the delta subunit to transfer the beta subunit on the DNA
- Gene Name:
- dnaX
- Locus Tag:
- PA1532
- Molecular weight:
- 73.3 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in RNA binding
- Specific function:
- Catalyzes the addition and repair of the essential 3'- terminal CCA sequence in tRNAs without using a nucleic acid template. Adds these three nucleotides in the order of C, C, and A to the tRNA nucleotide-73, using CTP and ATP as substrates and producing inorganic pyrophosphate. Also shows highest phosphatase activity in the presence of Ni(2+) and hydrolyzes pyrophosphate, canonical 5'-nucleoside tri- and diphosphates, NADP, and 2'-AMP with the production of Pi. Displays a metal-independent phosphodiesterase activity toward 2',3'-cAMP, 2',3'-cGMP, and 2',3'-cCMP. Without metal or in the presence of Mg(2+), this protein hydrolyzes 2',3'-cyclic substrates with the formation of 2'-nucleotides, whereas in the presence of Ni(2+), it also produces some 3'-nucleotides. These phosphohydrolase activities are probably involved in the repair of the tRNA 3'-CCA terminus degraded by intracellular RNases
- Gene Name:
- cca
- Locus Tag:
- PA0584
- Molecular weight:
- 45.6 kDa
Reactions
| A tRNA precursor + 2 CTP + ATP = a tRNA with a 3' CCA end + 3 diphosphate. |
- General function:
- Involved in hydrolase activity
- Specific function:
- This enzyme is involved in nucleotide metabolism:it produces dUMP, the immediate precursor of thymidine nucleotides and it decreases the intracellular concentration of dUTP so that uracil cannot be incorporated into DNA
- Gene Name:
- dut
- Locus Tag:
- PA5321
- Molecular weight:
- 15.9 kDa
Reactions
| dUTP + H(2)O = dUMP + diphosphate. |
- General function:
- Involved in phosphoribosyl-AMP cyclohydrolase activity
- Specific function:
- 1-(5-phosphoribosyl)-ATP + H(2)O = 1-(5- phosphoribosyl)-AMP + diphosphate
- Gene Name:
- hisI
- Locus Tag:
- PA5066
- Molecular weight:
- 15.4 kDa
Reactions
| 1-(5-phosphoribosyl)-ATP + H(2)O = 1-(5-phosphoribosyl)-AMP + diphosphate. |
| 1-(5-phosphoribosyl)-AMP + H(2)O = 1-(5-phosphoribosyl)-5-((5-phosphoribosylamino)methylideneamino)imidazole-4-carboxamide. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of valine to tRNA(Val). As ValRS can inadvertently accommodate and process structurally similar amino acids such as threonine, to avoid such errors, it has a "posttransfer" editing activity that hydrolyzes mischarged Thr-tRNA(Val) in a tRNA-dependent manner
- Gene Name:
- valS
- Locus Tag:
- PA3834
- Molecular weight:
- 107.7 kDa
Reactions
| ATP + L-valine + tRNA(Val) = AMP + diphosphate + L-valyl-tRNA(Val). |
- General function:
- Involved in RNA binding
- Specific function:
- ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe)
- Gene Name:
- pheT
- Locus Tag:
- PA2739
- Molecular weight:
- 86.8 kDa
Reactions
| ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-leucine + tRNA(Leu) = AMP + diphosphate + L-leucyl-tRNA(Leu)
- Gene Name:
- leuS
- Locus Tag:
- PA3987
- Molecular weight:
- 97.6 kDa
Reactions
| ATP + L-leucine + tRNA(Leu) = AMP + diphosphate + L-leucyl-tRNA(Leu). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe)
- Gene Name:
- pheS
- Locus Tag:
- PA2740
- Molecular weight:
- 38.1 kDa
Reactions
| ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe). |
- General function:
- Involved in metabolic process
- Specific function:
- Catalyzes the removal of elemental sulfur and selenium atoms from cysteine and selenocysteine to produce alanine. Functions as a sulfur delivery protein for NAD, biotin and Fe-S cluster synthesis. Transfers sulfur on 'Cys-456' of thiI in a transpersulfidation reaction. Transfers sulfur on 'Cys-19' of tusA in a transpersulfidation reaction. Functions also as a selenium delivery protein in the pathway for the biosynthesis of selenophosphate
- Gene Name:
- iscS
- Locus Tag:
- PA3814
- Molecular weight:
- 44.7 kDa
Reactions
| L-cysteine + acceptor = L-alanine + S-sulfanyl-acceptor. |
- General function:
- Involved in argininosuccinate synthase activity
- Specific function:
- ATP + L-citrulline + L-aspartate = AMP + diphosphate + N(omega)-(L-arginino)succinate
- Gene Name:
- argG
- Locus Tag:
- PA3525
- Molecular weight:
- 45.3 kDa
Reactions
| ATP + L-citrulline + L-aspartate = AMP + diphosphate + N(omega)-(L-arginino)succinate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Reversibly transfers an adenylyl group from ATP to 4'- phosphopantetheine, yielding dephospho-CoA (dPCoA) and pyrophosphate
- Gene Name:
- coaD
- Locus Tag:
- PA0363
- Molecular weight:
- 17.8 kDa
Reactions
| ATP + pantetheine 4'-phosphate = diphosphate + 3'-dephospho-CoA. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the reversible adenylation of nicotinate mononucleotide (NaMN) to nicotinic acid adenine dinucleotide (NaAD)
- Gene Name:
- nadD
- Locus Tag:
- PA4006
- Molecular weight:
- 23.8 kDa
Reactions
| ATP + beta-nicotinate-D-ribonucleotide = diphosphate + deamido-NAD(+). |
- General function:
- Involved in hydrolase activity
- Specific function:
- Master regulator of 5'-dependent mRNA decay. Accelerates the degradation of transcripts by removing pyrophosphate from the 5'-end of triphosphorylated RNA, leading to a more labile monophosphorylated state that can stimulate subsequent ribonuclease cleavage. Preferentially hydrolyzes diadenosine penta-phosphate with ATP as one of the reaction products. Also able to hydrolyze diadenosine hexa- and tetra-phosphate. Has no activity on diadenosine tri-phosphate, ADP-ribose, NADH and UDP- glucose. In the meningitis causing strain Pseudomonas aeruginosa K1, has been shown to play a role in HBMEC (human brain microvascular endothelial cells) invasion in vitro
- Gene Name:
- rppH
- Locus Tag:
- PA0336
- Molecular weight:
- 18.7 kDa
- General function:
- Involved in magnesium ion binding
- Specific function:
- Diphosphate + H(2)O = 2 phosphate
- Gene Name:
- ppa
- Locus Tag:
- PA4031
- Molecular weight:
- 19.4 kDa
Reactions
| Diphosphate + H(2)O = 2 phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the reversible transfer of the terminal phosphate of ATP to form a long-chain polyphosphate (polyP). Can form linear polymers of orthophosphate with chain lengths up to 1000 or more. Can also act in the reverse direction to form ATP in the presence of excess ADP. Can also use GTP instead of ATP; but the efficiency of GTP is 5% that of ATP
- Gene Name:
- ppk
- Locus Tag:
- PA5242
- Molecular weight:
- 83.2 kDa
Reactions
| ATP + (phosphate)(n) = ADP + (phosphate)(n+1). |
- General function:
- Involved in orotate phosphoribosyltransferase activity
- Specific function:
- Catalyzes the transfer of a ribosyl phosphate group from 5-phosphoribose 1-diphosphate to orotate, leading to the formation of orotidine monophosphate (OMP)
- Gene Name:
- pyrE
- Locus Tag:
- PA5331
- Molecular weight:
- 23.3 kDa
Reactions
| Orotidine 5'-phosphate + diphosphate = orotate + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in GTP cyclohydrolase II activity
- Specific function:
- Catalyzes the conversion of GTP to 2,5-diamino-6- ribosylamino-4(3H)-pyrimidinone 5'-phosphate (DARP), formate and pyrophosphate
- Gene Name:
- ribA
- Locus Tag:
- PA4047
- Molecular weight:
- 22.1 kDa
Reactions
| GTP + 3 H(2)O = formate + 2,5-diamino-6-hydroxy-4-(5-phospho-D-ribosylamino)pyrimidine + diphosphate. |
- General function:
- Involved in DNA-directed RNA polymerase activity
- Specific function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. This subunit plays an important role in subunit assembly since its dimerization is the first step in the sequential assembly of subunits to form the holoenzyme
- Gene Name:
- rpoA
- Locus Tag:
- PA4238
- Molecular weight:
- 36.6 kDa
Reactions
| Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1). |
- General function:
- Involved in DNA-directed RNA polymerase activity
- Specific function:
- Promotes RNA polymerase assembly. Latches the N- and C- terminal regions of the beta' subunit thereby facilitating its interaction with the beta and alpha subunits
- Gene Name:
- rpoZ
- Locus Tag:
- PA5337
- Molecular weight:
- 9.8 kDa
Reactions
| Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1). |
- General function:
- Involved in methionine adenosyltransferase activity
- Specific function:
- Catalyzes the formation of S-adenosylmethionine from methionine and ATP. The overall synthetic reaction is composed of two sequential steps, AdoMet formation and the subsequent tripolyphosphate hydrolysis which occurs prior to release of AdoMet from the enzyme. Is essential for growth
- Gene Name:
- metK
- Locus Tag:
- PA0546
- Molecular weight:
- 42.7 kDa
Reactions
| ATP + L-methionine + H(2)O = phosphate + diphosphate + S-adenosyl-L-methionine. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Nucleotidase with a broad substrate specificity as it can dephosphorylate various ribo- and deoxyribonucleoside 5'- monophosphates and ribonucleoside 3'-monophosphates with highest affinity to 3'-AMP. Also hydrolyzes polyphosphate (exopolyphosphatase activity) with the preference for short-chain- length substrates (P20-25). Might be involved in the regulation of dNTP and NTP pools, and in the turnover of 3'-mononucleotides produced by numerous intracellular RNases (T1, T2, and F) during the degradation of various RNAs. Also plays a significant physiological role in stress-response and is required for the survival of Pseudomonas aeruginosa in stationary growth phase
- Gene Name:
- surE
- Locus Tag:
- PA3625
- Molecular weight:
- 26.4 kDa
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| A 3'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| (Polyphosphate)(n) + H(2)O = (polyphosphate)(n-1) + phosphate. |
- General function:
- Involved in nucleoside metabolic process
- Specific function:
- Catalyzes the conversion of uracil and 5-phospho-alpha- D-ribose 1-diphosphate (PRPP) to UMP and diphosphate
- Gene Name:
- upp
- Locus Tag:
- PA4646
- Molecular weight:
- 22.9 kDa
Reactions
| UMP + diphosphate = uracil + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of serine to tRNA(Ser). Is also able to aminoacylate tRNA(Sec) with serine, to form the misacylated tRNA L-seryl-tRNA(Sec), which will be further converted into selenocysteinyl-tRNA(Sec)
- Gene Name:
- serS
- Locus Tag:
- PA2612
- Molecular weight:
- 47.2 kDa
Reactions
| ATP + L-serine + tRNA(Ser) = AMP + diphosphate + L-seryl-tRNA(Ser). |
| ATP + L-serine + tRNA(Sec) = AMP + diphosphate + L-seryl-tRNA(Sec). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ThrS is also a translational repressor protein, it controls the translation of its own gene by binding to its mRNA
- Gene Name:
- thrS
- Locus Tag:
- PA2744
- Molecular weight:
- 73.1 kDa
Reactions
| ATP + L-threonine + tRNA(Thr) = AMP + diphosphate + L-threonyl-tRNA(Thr). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-lysine + tRNA(Lys) = AMP + diphosphate + L-lysyl-tRNA(Lys)
- Gene Name:
- lysS
- Locus Tag:
- PA3700
- Molecular weight:
- 57.3 kDa
Reactions
| ATP + L-lysine + tRNA(Lys) = AMP + diphosphate + L-lysyl-tRNA(Lys). |
- General function:
- Involved in nucleotide binding
- Specific function:
- With YjeK might be involved in the post-translational modification of elongation factor P (EF-P)
- Gene Name:
- poxA
- Locus Tag:
- PA5513
- Molecular weight:
- 32.4 kDa
- General function:
- Involved in DNA binding
- Specific function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates
- Gene Name:
- rpoC
- Locus Tag:
- PA4269
- Molecular weight:
- 154.4 kDa
Reactions
| Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1). |
- General function:
- Involved in DNA binding
- Specific function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates
- Gene Name:
- rpoB
- Locus Tag:
- PA4270
- Molecular weight:
- 150.8 kDa
Reactions
| Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1). |
- General function:
- Involved in DNA binding
- Specific function:
- DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity. The beta chain is required for initiation of replication once it is clamped onto DNA, it slides freely (bidirectional and ATP- independent) along duplex DNA
- Gene Name:
- dnaN
- Locus Tag:
- PA0002
- Molecular weight:
- 40.7 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in RNA binding
- Specific function:
- Polymerase that creates the 3' poly(A) tail found in some mRNA's. Seems to be involved in plasmid copy number control
- Gene Name:
- pcnB
- Locus Tag:
- PA4727
- Molecular weight:
- 53.3 kDa
Reactions
| ATP + RNA(n) = diphosphate + RNA(n+1). |
- General function:
- Involved in transferase activity, transferring phosphorus-containing groups
- Specific function:
- CTP + phosphatidate = diphosphate + CDP- diacylglycerol
- Gene Name:
- cdsA
- Locus Tag:
- PA3651
- Molecular weight:
- 28.9 kDa
Reactions
| CTP + phosphatidate = diphosphate + CDP-diacylglycerol. |
- General function:
- Involved in phosphopantothenate--cysteine ligase activity
- Specific function:
- Catalyzes two steps in the biosynthesis of coenzyme A. In the first step cysteine is conjugated to 4'-phosphopantothenate to form 4-phosphopantothenoylcysteine, in the latter compound is decarboxylated to form 4'-phosphopantotheine
- Gene Name:
- coaBC
- Locus Tag:
- PA5320
- Molecular weight:
- 43.1 kDa
Reactions
| N-((R)-4'-phosphopantothenoyl)-L-cysteine = pantotheine 4'-phosphate + CO(2). |
| CTP + (R)-4'-phosphopantothenate + L-cysteine = CMP + diphosphate + N-((R)-4'-phosphopantothenoyl)-L-cysteine. |
- General function:
- Involved in dihydropteroate synthase activity
- Specific function:
- DHPS catalyzes the formation of the immediate precursor of folic acid. It is implicated in resistance to sulfonamide
- Gene Name:
- folP
- Locus Tag:
- PA4750
- Molecular weight:
- 30.5 kDa
Reactions
| (2-amino-4-hydroxy-7,8-dihydropteridin-6-yl)methyl diphosphate + 4-aminobenzoate = diphosphate + dihydropteroate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the last two sequential reactions in the de novo biosynthetic pathway for UDP-GlcNAc. Responsible for the acetylation of Glc-N-1-P to give GlcNAc-1-P and for the uridyl transfer from UTP to GlcNAc-1-P which produces UDP-GlcNAc
- Gene Name:
- glmU
- Locus Tag:
- PA5552
- Molecular weight:
- 48.9 kDa
Reactions
| Acetyl-CoA + alpha-D-glucosamine 1-phosphate = CoA + N-acetyl-alpha-D-glucosamine 1-phosphate. |
| UTP + N-acetyl-alpha-D-glucosamine 1-phosphate = diphosphate + UDP-N-acetyl-D-glucosamine. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP-dependent phosphorylation of adenosylcobinamide and adds GMP to adenosylcobinamide phosphate
- Gene Name:
- cobU
- Locus Tag:
- PA1279
- Molecular weight:
- 36.5 kDa
Reactions
| ATP or GTP + adenosylcobinamide = adenosylcobinamide phosphate + ADP or GDP. |
| GTP + adenosylcobinamide phosphate = diphosphate + adenosylcobinamide-GDP. |
- General function:
- Involved in UTP:glucose-1-phosphate uridylyltransferase activity
- Specific function:
- May play a role in stationary phase survival
- Gene Name:
- galU
- Locus Tag:
- PA2023
- Molecular weight:
- 31.2 kDa
Reactions
| UTP + alpha-D-glucose 1-phosphate = diphosphate + UDP-glucose. |
- General function:
- Involved in nucleoside-triphosphate diphosphatase activity
- Specific function:
- Specific function unknown
- Gene Name:
- mazG
- Locus Tag:
- PA0935
- Molecular weight:
- 31.2 kDa
Reactions
| ATP + H(2)O = AMP + diphosphate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Degradation of inorganic polyphosphates. Orthophosphate is released progressively from the ends of polyphosphate of circa 500 residues long, while chains of circa 15 residues compete poorly with polyphosphate as substrate
- Gene Name:
- ppx
- Locus Tag:
- PA5241
- Molecular weight:
- 56.4 kDa
Reactions
| (Polyphosphate)(n) + H(2)O = (polyphosphate)(n-1) + phosphate. |
- General function:
- Involved in amidophosphoribosyltransferase activity
- Specific function:
- 5-phospho-beta-D-ribosylamine + diphosphate + L-glutamate = L-glutamine + 5-phospho-alpha-D-ribose 1-diphosphate + H(2)O
- Gene Name:
- purF
- Locus Tag:
- PA3108
- Molecular weight:
- 55.4 kDa
Reactions
| 5-phospho-beta-D-ribosylamine + diphosphate + L-glutamate = L-glutamine + 5-phospho-alpha-D-ribose 1-diphosphate + H(2)O. |
- General function:
- Involved in catalytic activity
- Specific function:
- In eubacteria ppGpp (guanosine 3'-diphosphate 5-' diphosphate) is a mediator of the stringent response that coordinates a variety of cellular activities in response to changes in nutritional abundance. This enzyme catalyzes both the synthesis and degradation of ppGpp. The second messengers ppGpp and c-di-GMP together control biofilm formation in response to translational stress; ppGpp represses biofilm formation while c- di-GMP induces it
- Gene Name:
- spoT
- Locus Tag:
- PA5338
- Molecular weight:
- 78.9 kDa
Reactions
| ATP + GTP = AMP + guanosine 3'-diphosphate 5'-triphosphate. |
| Guanosine 3',5'-bis(diphosphate) + H(2)O = guanosine 5'-diphosphate + diphosphate. |
- General function:
- Involved in FMN adenylyltransferase activity
- Specific function:
- ATP + riboflavin = ADP + FMN
- Gene Name:
- ribF
- Locus Tag:
- PA4561
- Molecular weight:
- 34.3 kDa
Reactions
| ATP + riboflavin = ADP + FMN. |
| ATP + FMN = diphosphate + FAD. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of tyrosine to tRNA(Tyr) in a two-step reaction:tyrosine is first activated by ATP to form Tyr- AMP and then transferred to the acceptor end of tRNA(Tyr)
- Gene Name:
- tyrS
- Locus Tag:
- PA4138
- Molecular weight:
- 45.1 kDa
Reactions
| ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr). |
- General function:
- Involved in 3'-5' exonuclease activity
- Specific function:
- DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity. The alpha chain is the DNA polymerase
- Gene Name:
- dnaE
- Locus Tag:
- PA3640
- Molecular weight:
- 130.9 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-arginine + tRNA(Arg) = AMP + diphosphate + L-arginyl-tRNA(Arg)
- Gene Name:
- argS
- Locus Tag:
- PA5051
- Molecular weight:
- 65.2 kDa
Reactions
| ATP + L-arginine + tRNA(Arg) = AMP + diphosphate + L-arginyl-tRNA(Arg). |
- General function:
- Involved in ATP binding
- Specific function:
- Catalyzes the transfer of a dimethylallyl group onto the adenine at position 37 in tRNAs that read codons beginning with uridine, leading to the formation of N6-(dimethylallyl)adenosine (i(6)A)
- Gene Name:
- miaA
- Locus Tag:
- PA4945
- Molecular weight:
- 35.8 kDa
Reactions
| Dimethylallyl diphosphate + tRNA = diphosphate + tRNA containing 6-dimethylallyladenosine. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of proline to tRNA(Pro) in a two-step reaction:proline is first activated by ATP to form Pro- AMP and then transferred to the acceptor end of tRNA(Pro). As ProRS can inadvertently accommodate and process non-cognate amino acids such as alanine and cysteine, to avoid such errors it has two additional distinct editing activities against alanine. One activity is designated as 'pretransfer' editing and involves the tRNA(Pro)-independent hydrolysis of activated Ala-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Ala-tRNA(Pro). Misacylated Cys-tRNA(Pro) is not edited by ProRS, but instead may be edited in trans by ybaK
- Gene Name:
- proS
- Locus Tag:
- PA0956
- Molecular weight:
- 63.1 kDa
Reactions
| ATP + L-proline + tRNA(Pro) = AMP + diphosphate + L-prolyl-tRNA(Pro). |
- General function:
- Involved in nicotinate phosphoribosyltransferase activity
- Specific function:
- Nicotinate D-ribonucleotide + diphosphate = nicotinate + 5-phospho-alpha-D-ribose 1-diphosphate
- Gene Name:
- pncB
- Locus Tag:
- PA4919
- Molecular weight:
- 46.1 kDa
Reactions
| Beta-nicotinate D-ribonucleotide + diphosphate = nicotinate + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in NAD+ synthase (glutamine-hydrolyzing) activity
- Specific function:
- Catalyzes a key step in NAD biosynthesis, transforming deamido-NAD into NAD by a two-step reaction
- Gene Name:
- nadE
- Locus Tag:
- PA4920
- Molecular weight:
- 29.7 kDa
Reactions
| ATP + deamido-NAD(+) + NH(3) = AMP + diphosphate + NAD(+). |
- General function:
- Involved in catalytic activity
- Specific function:
- ATP + sulfate = diphosphate + adenylyl sulfate
- Gene Name:
- cysD
- Locus Tag:
- PA4443
- Molecular weight:
- 35.5 kDa
Reactions
| ATP + sulfate = diphosphate + adenylyl sulfate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Thought to be involved in DNA repair and/or mutagenesis
- Gene Name:
- polB
- Locus Tag:
- PA1886
- Molecular weight:
- 89.7 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in cysteine-tRNA ligase activity
- Specific function:
- ATP + L-cysteine + tRNA(Cys) = AMP + diphosphate + L-cysteinyl-tRNA(Cys)
- Gene Name:
- cysS
- Locus Tag:
- PA1795
- Molecular weight:
- 51.2 kDa
Reactions
| ATP + L-cysteine + tRNA(Cys) = AMP + diphosphate + L-cysteinyl-tRNA(Cys). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-aspartate + tRNA(Asp) = AMP + diphosphate + L-aspartyl-tRNA(Asp)
- Gene Name:
- aspS
- Locus Tag:
- PA0963
- Molecular weight:
- 66.2 kDa
Reactions
| ATP + L-aspartate + tRNA(Asp) = AMP + diphosphate + L-aspartyl-tRNA(Asp). |
- General function:
- Involved in isoprenoid biosynthetic process
- Specific function:
- Geranyl diphosphate + isopentenyl diphosphate = diphosphate + trans,trans-farnesyl diphosphate
- Gene Name:
- ispA
- Locus Tag:
- PA4043
- Molecular weight:
- 31.5 kDa
Reactions
| Geranyl diphosphate + isopentenyl diphosphate = diphosphate + (2E,6E)-farnesyl diphosphate. |
- General function:
- Involved in GTPase activity
- Specific function:
- May be the GTPase, regulating ATP sulfurylase activity
- Gene Name:
- cysN
- Locus Tag:
- PA4442
- Molecular weight:
- 69.3 kDa
Reactions
| ATP + sulfate = diphosphate + adenylyl sulfate. |
- General function:
- Involved in nucleotidyltransferase activity
- Specific function:
- Involved in the biosynthesis of the capsular polysaccharide colanic acid
- Gene Name:
- manC
- Locus Tag:
- PA3551
- Molecular weight:
- 53.1 kDa
Reactions
| GTP + alpha-D-mannose 1-phosphate = diphosphate + GDP-mannose. |
- General function:
- Involved in sulfurtransferase activity
- Specific function:
- Catalyzes the 2-thiolation of uridine at the wobble position (U34) of tRNA(Lys), tRNA(Glu) and tRNA(Gln), leading to the formation of s(2)U34, the first step of tRNA-mnm(5)s(2)U34 synthesis. Sulfur is provided by iscS, via a sulfur-relay system. Binds ATP and its substrate tRNAs
- Gene Name:
- mnmA
- Locus Tag:
- PA2626
- Molecular weight:
- 41.9 kDa
- General function:
- Involved in amino acid binding
- Specific function:
- Modifies, by uridylylation or deuridylylation the PII (glnB) regulatory protein
- Gene Name:
- glnD
- Locus Tag:
- PA3658
- Molecular weight:
- 103.4 kDa
Reactions
| UTP + [protein-PII] = diphosphate + uridylyl-[protein-PII]. |
- General function:
- Involved in acetate-CoA ligase activity
- Specific function:
- Enables the cell to use acetate during aerobic growth to generate energy via the TCA cycle, and biosynthetic compounds via the glyoxylate shunt. Acetylates CheY, the response regulator involved in flagellar movement and chemotaxis
- Gene Name:
- acs
- Locus Tag:
- PA0887
- Molecular weight:
- 71.8 kDa
Reactions
| ATP + acetate + CoA = AMP + diphosphate + acetyl-CoA. |
- General function:
- Involved in DNA binding
- Specific function:
- DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity. The delta subunit seems to interact with the gamma subunit to transfer the beta subunit on the DNA
- Gene Name:
- holA
- Locus Tag:
- PA3989
- Molecular weight:
- 37.4 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in DNA binding
- Specific function:
- DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity
- Gene Name:
- holB
- Locus Tag:
- PA2961
- Molecular weight:
- 35.7 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in DNA binding
- Specific function:
- DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity
- Gene Name:
- holC
- Locus Tag:
- PA3832
- Molecular weight:
- 16.1 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in catalytic activity
- Specific function:
- Involved in the catabolism of quinolinic acid (QA)
- Gene Name:
- nadC
- Locus Tag:
- PA4524
- Molecular weight:
- 30.6 kDa
Reactions
| Beta-nicotinate D-ribonucleotide + diphosphate + CO(2) = pyridine-2,3-dicarboxylate + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Condenses 4-methyl-5-(beta-hydroxyethyl)thiazole monophosphate (THZ-P) and 2-methyl-4-amino-5-hydroxymethyl pyrimidine pyrophosphate (HMP-PP) to form thiamine monophosphate (TMP)
- Gene Name:
- thiE
- Locus Tag:
- PA3976
- Molecular weight:
- 22.1 kDa
Reactions
| 2-methyl-4-amino-5-hydroxymethylpyrimidine diphosphate + 4-methyl-5-(2-phosphono-oxyethyl)thiazole = diphosphate + thiamine phosphate. |
- General function:
- Involved in [glutamate-ammonia-ligase] adenylyltransferase activity
- Specific function:
- Adenylation and deadenylation of glutamate--ammonia ligase
- Gene Name:
- glnE
- Locus Tag:
- PA5014
- Molecular weight:
- 110.1 kDa
Reactions
| ATP + [L-glutamate:ammonia ligase (ADP-forming)] = diphosphate + adenylyl-[L-glutamate:ammonia ligase (ADP-forming)]. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the condensation of pantoate with beta-alanine in an ATP-dependent reaction via a pantoyl-adenylate intermediate
- Gene Name:
- panC
- Locus Tag:
- PA4730
- Molecular weight:
- 30.8 kDa
Reactions
| ATP + (R)-pantoate + beta-alanine = AMP + diphosphate + (R)-pantothenate. |
- General function:
- Involved in ATP binding
- Specific function:
- Catalyzes two reactions:the first one is the production of beta-formyl glycinamide ribonucleotide (GAR) from formate, ATP and beta GAR; the second, a side reaction, is the production of acetyl phosphate and ADP from acetate and ATP
- Gene Name:
- purT
- Locus Tag:
- PA3751
- Molecular weight:
- 42.3 kDa
Reactions
| Formate + ATP + 5'-phospho-ribosylglycinamide = 5'-phosphoribosyl-N-formylglycinamide + ADP + diphosphate. |
- General function:
- Involved in glucose-1-phosphate thymidylyltransferase activity
- Specific function:
- Catalyzes the formation of dTDP-glucose, from dTTP and glucose 1-phosphate, as well as its pyrophosphorolysis
- Gene Name:
- rmlA1
- Locus Tag:
- PA5163
- Molecular weight:
- 32.5 kDa
Reactions
| dTTP + alpha-D-glucose 1-phosphate = diphosphate + dTDP-glucose. |
- General function:
- Involved in RNA processing
- Specific function:
- Catalyzes the conversion of 3'-phosphate to a 2',3'- cyclic phosphodiester at the end of RNA. The mechanism of action of the enzyme occurs in 3 steps:(A) adenylation of the enzyme by ATP; (B) the enzyme acts on RNA-N3'P to produce RNA-N3'PP5'A; (C) a non catalytic nucleophilic attack by the adjacent 2'hydroxyl on the phosphorus in the diester linkage to produce the cyclic end product. The biological role of this enzyme is unknown but it is likely to function in some aspects of cellular RNA processing
- Gene Name:
- rtcA
- Locus Tag:
- PA4585
- Molecular weight:
- 36.6 kDa
Reactions
| ATP + RNA 3'-terminal-phosphate = AMP + diphosphate + RNA terminal-2',3'-cyclic-phosphate. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Hydrolyzes O6 atom-containing purine bases deoxyinosine triphosphate (dITP) and xanthosine triphosphate (XTP) as well as 2'-deoxy-N-6-hydroxylaminopurine triposphate (dHAPTP) to nucleotide monophosphate and pyrophosphate. Probably excludes non- standard purines from DNA precursor pool, preventing thus incorporation into DNA and avoiding chromosomal lesions
- Gene Name:
- rdgB
- Locus Tag:
- PA0387
- Molecular weight:
- 21.2 kDa
Reactions
| A nucleoside triphosphate + H(2)O = a nucleotide + diphosphate. |
- General function:
- Involved in transferase activity, transferring alkyl or aryl (other than methyl) groups
- Specific function:
- Generates undecaprenyl pyrophosphate (UPP) from isopentenyl pyrophosphate (IPP). UPP is the precursor of glycosyl carrier lipid in the biosynthesis of bacterial cell wall polysaccharide components such as peptidoglycan and lipopolysaccharide
- Gene Name:
- uppS
- Locus Tag:
- PA3652
- Molecular weight:
- 28 kDa
Reactions
| (2E,6E)-farnesyl diphosphate + 8 isopentenyl diphosphate = 8 diphosphate + di-trans,octa-cis-undecaprenyl diphosphate. |
- General function:
- Involved in ATP phosphoribosyltransferase activity
- Specific function:
- Catalyzes the condensation of ATP and 5-phosphoribose 1- diphosphate to form N'-(5'-phosphoribosyl)-ATP (PR-ATP). Has a crucial role in the pathway because the rate of histidine biosynthesis seems to be controlled primarily by regulation of hisG enzymatic activity
- Gene Name:
- hisG
- Locus Tag:
- PA4449
- Molecular weight:
- 22.8 kDa
Reactions
| 1-(5-phospho-D-ribosyl)-ATP + diphosphate = ATP + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-histidine + tRNA(His) = AMP + diphosphate + L-histidyl-tRNA(His)
- Gene Name:
- hisS
- Locus Tag:
- PA3802
- Molecular weight:
- 47.5 kDa
Reactions
| ATP + L-histidine + tRNA(His) = AMP + diphosphate + L-histidyl-tRNA(His). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the esterification, concomitant with transport, of exogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids
- Gene Name:
- fadD
- Locus Tag:
- PA3299
- Molecular weight:
- 61.7 kDa
Reactions
| ATP + a long-chain fatty acid + CoA = AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in adenine phosphoribosyltransferase activity
- Specific function:
- Catalyzes a salvage reaction resulting in the formation of AMP, that is energically less costly than de novo synthesis
- Gene Name:
- apt
- Locus Tag:
- PA1543
- Molecular weight:
- 19.8 kDa
Reactions
| AMP + diphosphate = adenine + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in damaged DNA binding
- Specific function:
- Poorly processive, error-prone DNA polymerase involved in untargeted mutagenesis. Copies undamaged DNA at stalled replication forks, which arise in vivo from mismatched or misaligned primer ends. These misaligned primers can be extended by polIV. Exhibits no 3'-5' exonuclease (proofreading) activity. Overexpression of polIV results in increased frameshift mutagenesis. It is required for stationary-phase adaptive mutation, which provides the bacterium with flexibility in dealing with environmental stress, enhancing long-term survival and evolutionary fitness. May be involved in translesional synthesis, in conjunction with the beta clamp from polIII
- Gene Name:
- dinB
- Locus Tag:
- PA0923
- Molecular weight:
- 38.9 kDa
Reactions
| Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- Catalyzes the ADP transfer to D-glycero-D-manno-heptose 1-phosphate, yielding ADP-D,D-heptose
- Gene Name:
- hldE
- Locus Tag:
- PA4996
- Molecular weight:
- 50.3 kDa
Reactions
| ATP + D-glycero-beta-D-manno-heptose 7-phosphate = ADP + D-glycero-beta-D-manno-heptose 1,7-bisphosphate. |
| ATP + D-glycero-beta-D-manno-heptose 1-phosphate = diphosphate + ADP-D-glycero-beta-D-manno-heptose. |
- General function:
- Involved in catalytic activity
- Specific function:
- Links a guanosine 5'-phosphate to molydopterin (MPT) forming molybdopterin guanine dinucleotide (MGD)
- Gene Name:
- mobA
- Locus Tag:
- PA3030
- Molecular weight:
- 21.9 kDa
Reactions
| GTP + molybdenum cofactor = diphosphate + guanylyl molybdenum cofactor. |
- General function:
- Involved in prenyltransferase activity
- Specific function:
- Synthesis of 3-octaprenyl-4-hydroxybenzoate
- Gene Name:
- ubiA
- Locus Tag:
- PA5358
- Molecular weight:
- 33 kDa
Reactions
| 4-hydroxybenzoate + farnesylfarnesylgeraniol = 3-octaprenyl-4-hydroxybenzoate. |
- General function:
- Involved in RNA binding
- Specific function:
- Catalyzes the ATP-dependent transfer of a sulfur to tRNA to produce 4-thiouridine in position 8 of tRNAs, which functions as a near-UV photosensor. Also catalyzes the transfer of sulfur to the sulfur carrier protein ThiS, forming ThiS-thiocarboxylate. This is a step in the synthesis of thiazole, in the thiamine biosynthesis pathway. The sulfur is donated as persulfide by iscS
- Gene Name:
- thiI
- Locus Tag:
- PA5118
- Molecular weight:
- 54.8 kDa
Reactions
| L-cysteine + 'activated' tRNA = L-serine + tRNA containing a thionucleotide. |
| [IscS]-SSH + [ThiS]-COAMP = [IscS]-SH + [ThiS]-COSH + AMP. |
- General function:
- Involved in isoprenoid biosynthetic process
- Specific function:
- Supplies octaprenyl diphosphate, the precursor for the side chain of the isoprenoid quinones ubiquinone and menaquinone
- Gene Name:
- ispB
- Locus Tag:
- PA4569
- Molecular weight:
- 34.9 kDa
Reactions
| (2E,6E)-farnesyl diphosphate + 5 isopentenyl diphosphate = 5 diphosphate + all-trans-octaprenyl diphosphate. |
- General function:
- Involved in protoheme IX farnesyltransferase activity
- Specific function:
- Converts heme B (protoheme IX) to heme O by substitution of the vinyl group on carbon 2 of heme B porphyrin ring with a hydroxyethyl farnesyl side group
- Gene Name:
- cyoE
- Locus Tag:
- PA1321
- Molecular weight:
- 31.8 kDa
- General function:
- Involved in Mo-molybdopterin cofactor biosynthetic process
- Specific function:
- Catalyzes the adenylation by ATP of the carboxyl group of the C-terminal glycine of sulfur carrier protein moaD
- Gene Name:
- moeB
- Locus Tag:
- PA4663
- Molecular weight:
- 26.7 kDa
Reactions
| ATP + [molybdopterin-synthase sulfur-carrier protein]-Gly-Gly = diphosphate + [molybdopterin-synthase sulfur-carrier protein]-Gly-Gly-AMP. |
- General function:
- Involved in Mo-molybdopterin cofactor biosynthetic process
- Specific function:
- Together with moaA, is involved in the conversion of a guanosine derivative (5'-GTP) into molybdopterin precursor Z
- Gene Name:
- moaC
- Locus Tag:
- PA3918
- Molecular weight:
- 17.3 kDa
- General function:
- Involved in fatty acid biosynthetic process
- Specific function:
- Carrier of the growing fatty acid chain in fatty acid biosynthesis
- Gene Name:
- acpP
- Locus Tag:
- PA2966
- Molecular weight:
- 8.7 kDa
- General function:
- Replication, recombination and repair
- Specific function:
- Involved in the GO system responsible for removing an oxidatively damaged form of guanine (7,8-dihydro-8-oxoguanine) from DNA and the nucleotide pool. 8-oxo-dGTP is inserted opposite dA and dC residues of template DNA with almost equal efficiency thus leading to A.T to G.C transversions. MutT specifically degrades 8-oxo-dGTP to the monophosphate
- Gene Name:
- mutT
- Locus Tag:
- PA4400
- Molecular weight:
- 34 kDa
Reactions
| 8-oxo-dGTP + H(2)O = 8-oxo-dGMP + diphosphate. |
- General function:
- threonylcarbamoyladenosine biosynthetic process
- Specific function:
- Required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. Catalyzes the conversion of L-threonine, bicarbonate/CO(2) and ATP to give threonylcarbamoyl-AMP (TC-AMP) as the acyladenylate intermediate, with the release of pyrophosphate. Is also able to catalyze the reverse reaction in vitro, i.e. the formation of ATP from TC-AMP and PPi. Shows higher affinity for the full-length tRNA(Thr) lacking only the t(6)A37 modification than for its fully modified counterpart. Could also be required for the maturation of 16S rRNA. Binds to double-stranded RNA but does not interact tightly with either of the ribosomal subunits, or the 70S particles.
- Gene Name:
- tsaC
- Locus Tag:
- PA0022
- Molecular weight:
- 20.4 kDa
Reactions
| L-threonine + ATP + bicarbonate = L-threonylcarbamoyladenylate + diphosphate + H(2)O |
- General function:
- organic phosphonate catabolic process
- Specific function:
- Catalyzes the hydrolysis of alpha-D-ribose 1-methylphosphonate triphosphate (RPnTP) to form alpha-D-ribose 1-methylphosphonate phosphate (PRPn) and diphosphate.
- Gene Name:
- phnM
- Locus Tag:
- PA3374
- Molecular weight:
- 41.6 kDa
Reactions
| Alpha-D-ribose 1-methylphosphonate 5-triphosphate + H(2)O = alpha-D-ribose 1-methylphosphonate 5-phosphate + diphosphate |
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for phosphonates; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- phnE
- Locus Tag:
- PA3382
- Molecular weight:
- 28.4 kDa