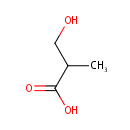
Search Results for compounds
Searching compounds for
returned 4373 results.
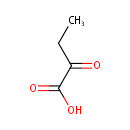
2-Ketobutyric acid (PAMDB000001)
IUPAC:
2-oxobutanoic acid
CAS: 600-18-0
Description: 2-Ketobutyric acid (alpha ketobutyric acid) is involved in the metabolism of many amino acids (glycine, cysteine, methionine, valine, leucine, serine, threonine, isoleucine). It also plays a role in propanoate metabolism and C-5 branched dibasic acid metabolism. More specifically, alpha-ketobutyric acid can be produced through the lysis of cystathionine (via cystathionine gamma lyase) leading to the production of cysteine and alpha-ketobutyric acid. It is also one of the degradation products of threonine. It can be converted to propionyl-CoA (and subsequently methylmalonyl CoA, which can be converted to succinyl CoA, a citric acid cycle intermediate), and thus enter the citric acid cycle.
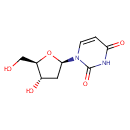
Deoxyuridine (PAMDB000002)
IUPAC:
1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,3,4-tetrahydropyrimidine-2,4-dione
CAS: 951-78-0
Description: 2'-Deoxyuridine is a naturally occurring nucleoside. It is similar in chemical structure to uridine, but without the 2'-hydroxyl group. It is considered to be an antimetabolite that is converted to deoxyuridine triphosphate during DNA synthesis.
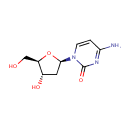
Deoxycytidine (PAMDB000003)
IUPAC:
4-amino-1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2-dihydropyrimidin-2-one
CAS: 951-77-9
Description: Deoxycytidine is one of the principal nucleosides of DNA composed of cytosine and deoxyribose. A nucleoside consists of only a pentose sugar linked to a purine or pyrimidine base, without a phosphate group. When N1 is linked to the C1 of deoxyribose, deoxynucleosides and nucleotides are formed from cytosine and deoxyribose; deoxycytidine monophosphate (dCMP), deoxycytidine diphosphate (dCDP), deoxycytidine triphosphate (dCTP). CTP is the source of the cytidine in RNA (ribonucleic acid) and deoxycytidine triphosphate (dCTP) is the source of the deoxycytidine in DNA (deoxyribonucleic acid).
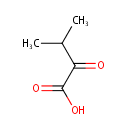
a-Ketoisovaleric acid (PAMDB000004)
IUPAC:
3-methyl-2-oxobutanoic acid
CAS: 759-05-7
Description: a-Ketoisovaleric acid is a precursor of valine, leucine and pantothenate. It can be synthesized in Pseudomonas aeruginosa. anabolically (via dihydroxyacid dehydrase) and catabolically (branched-chain amino acid transaminase and alanine-valine transaminase)
3-Hydroxyisobutyric acid (PAMDB000005)
IUPAC:
3-hydroxy-2-methylpropanoic acid
CAS: 2068-83-9
Description: 3-Hydroxyisobutyric acid is a member of the chemical class known as Beta Hydroxy Acids and Derivatives. These are compounds containing a carboxylic acid substituted with a hydroxyl group on the C3 carbon atom. 3-Hydroxyisobutyric acid (or 3-hydroxy-2-methylpropanoic acid) is an intermediate in the metabolism of valine. (WikiPedia)
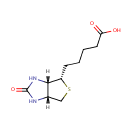
Biotin (PAMDB000008)
IUPAC:
5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4-d]imidazolidin-4-yl]pentanoic acid
CAS: 58-85-5
Description: Biotin is an enzyme co-factor present in minute amounts in every living cell. Biotin is also known as vitamin H or B7 or coenzyme R. It occurs mainly bound to proteins or polypeptides. The utilization of biotin for covalent attachment to carboxylases and its reutilization through the release of carboxylase biotin after proteolytic degradation constitutes the 'biotin cycle'. Biotin acts as a carboxyl carrier in carboxylation reactions. The biotin moiety is covalently bound to the epsilon amino group of a Lysine residue in each of these carboxylases in a domain 60-80 amino acids long. The domain is structurally similar among carboxylases from bacteria to mammals. The biotin moiety is covalently bound to the epsilon amino group of a Lys residue in carboxylases in a domain 60-80 amino acids long. The domain is structurally similar among carboxylases from bacteria to mammals. Posttranscriptional events related to ribosomal activity and protein folding may further contribute to effects of biotin on gene expression.
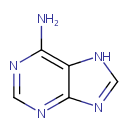
Adenine (PAMDB000010)
IUPAC:
7H-purin-6-amine
CAS: 73-24-5
Description: Adenine is a purine base. Adenine is found in both DNA and RNA. Adenine is a fundamental component of adenine nucleotides. Adenine forms adenosine, a nucleoside, when attached to ribose, and deoxyadenosine when attached to deoxyribose; it forms adenosine triphosphate (ATP), a nucleotide, when three phosphate groups are added to adenosine. Adenosine triphosphate is used in cellular metabolism as one of the basic methods of transferring chemical energy between chemical reactions.
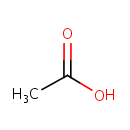
Acetic acid (PAMDB000012)
IUPAC:
acetic acid
CAS: 64-19-7
Description: Acetic acid is one of the simplest carboxylic acids. The acetyl group, derived from acetic acid, is fundamental to the biochemistry of virtually all forms of life. When bound to coenzyme A it forms acetyl-CoA, which is central to the metabolism of carbohydrates and fats. However, the concentration of free acetic acid in cells is kept at a low level to avoid disrupting the control of the pH of the cell contents. Acetic acid is produced and excreted by certain bacteria, notably the Acetobacter genus and Clostridium acetobutylicum. These bacteria are found universally in foodstuffs, water, and soil, and acetic acid is produced naturally as fruits and some other foods spoil. (Wikipedia) Acetic acid, or more accurately, acetate, a derivative of acetic acid, can be produced by Pseudomonas aeruginosa through fermentation in glucose metabolism. (KEGG, PMID 18600996)
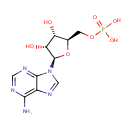
Adenosine monophosphate (PAMDB000014)
IUPAC:
{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid
CAS: 61-19-8
Description: Adenosine monophosphate, also known as 5'-adenylic acid and abbreviated AMP, is a nucleotide that is found in RNA. It is an ester of phosphoric acid with the nucleoside adenosine. AMP consists of the phosphate group, the pentose sugar ribose, and the nucleobase adenine. AMP can be produced during ATP synthesis by the enzyme adenylate kinase. AMP has recently been approved as a 'Bitter Blocker' additive to foodstuffs. When AMP is added to bitter foods or foods with a bitter aftertaste it makes them seem 'sweeter'. This potentially makes lower calorie food products more palatable
Melibiose (PAMDB000015)
IUPAC:
(3R,4S,5S,6R)-6-({[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}methyl)oxane-2,3,4,5-tetrol
CAS: 585-99-9
Description: Melibiose is a disaccharide consisting of one galactose and one glucose moiety in an alpha (1-6) glycosidic linkage. This sugar is produced and metabolized only by enteric and lactic acid bacteria and other microbes. Melibiose can be broken down by gut microflora such as Pseudomonas aeruginosa. Melibiose is first imported by the melibiose permease, MelB, and then converted to alpha-D-glucose and alpha-D-galactose by the galactosidase encoded by melA. (HMDB) In fact, Pseudomonas aeruginosa is able to utilize melibiose as a sole source of carbon.