| Identification |
| Name: |
Phosphoribosylglycinamide formyltransferase 2 |
| Synonyms: |
- GART 2
- 5'-phosphoribosylglycinamide transformylase 2
- Formate-dependent GAR transformylase
- GAR transformylase 2
|
| Gene Name: |
purT |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in ATP binding |
| Specific Function: |
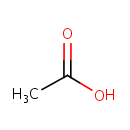
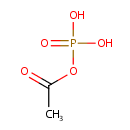
Catalyzes two reactions:the first one is the production of beta-formyl glycinamide ribonucleotide (GAR) from formate, ATP and beta GAR; the second, a side reaction, is the production of acetyl phosphate and ADP from acetate and ATP |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
Tetrahydrofolic acid | + | 5'-Phosphoribosyl-N-formylglycinamide | + | 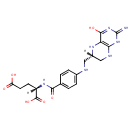 | + | 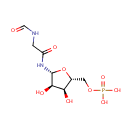 | → |  | + |  | + | Glycineamideribotide | + | 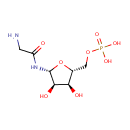 |
| | |
Tetrahydrofolic acid | + | 5'-Phosphoribosyl-N-formylglycinamide | + |  | + |  | → | 10-Formyltetrahydrofolate | + | Glycineamideribotide | + |  | + |  |
| | |
 | + | Tetrahydrofolic acid | + |  | → |  | + | Glycineamideribotide | + |  | + |  |
| | |
 | + |  | + | 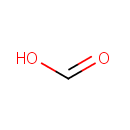 | → | 5'-Phosphoribosyl-N-formylglycinamide | + | Adenosine diphosphate | + | 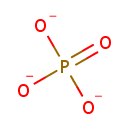 | + |  | + |  | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | catalytic activity | | cation binding | | glycine hydroxymethyltransferase activity | | hydroxymethyl-, formyl- and related transferase activity | | ion binding | | magnesium ion binding | | metal ion binding | | methyltransferase activity | | nucleoside binding | | purine nucleoside binding | | transferase activity | | transferase activity, transferring one-carbon groups | | Process |
|---|
| cellular nitrogen compound metabolic process | | metabolic process | | nitrogen compound metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleoside phosphate metabolic process | | nucleotide metabolic process | | purine nucleotide biosynthetic process | | purine nucleotide metabolic process | | purine ribonucleotide biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA3751 |
| Strand: |
- |
| Entrez Gene ID: |
880455 |
| Accession: |
NP_252440.1 |
| GI: |
15598946 |
| Sequence start: |
4203324 |
| Sequence End: |
4204505 |
| Sequence Length: |
1181 |
| Gene Sequence: |
>PA3751
ATGACCCGTATCGGTACCCCCCTGTCGCCGAGCGCGACCCGCGTCCTTCTCTGTGGCTCCGGCGAGCTGGGCAAGGAAGTCGCCATCGAGCTGCAACGCCTGGGTTGCGAAGTGATCGCCGTCGATCGCTATGGCAACGCGCCGGCCATGCAGGTGGCACATCGCAGCCACGTGATCAGCATGCTCGACGGCGCCGCCCTGCGCGCGGTGATCGAGCAGGAGAAGCCGCACTACATCGTGCCGGAGATCGAGGCCATCGCCACCGCGACCCTGGTCGAGCTGGAGGCCGAAGGCTACACCGTGGTGCCCACCGCCCGCGCCGCGCAACTGACCATGAACCGCGAGGGTATCCGCCGGCTGGCTGCCGAGGAGCTCGGCTTGCCGACCTCGCCCTATCACTTCGCCGATACCTTCGAGGACTACCGCCGTGGCGTCGAGCGCGTCGGCTACCCCTGCGTGGTGAAGCCGATCATGAGTTCGTCGGGCAAGGGCCAGAGCGTCCTGAAAGGCCCCGACGACCTGCAGGCGGCCTGGGACTATGCCCAGGAAGGCGGGCGCGCCGGCAAGGGCCGGGTGATCGTCGAGGGCTTCATCGATTTCGACTACGAAATCACCCTGCTCACCGTGCGCCACGTCGACGGCACCACTTTCTGCGCGCCGATCGGTCACCGCCAGGTCAAGGGCGACTACCACGAGTCCTGGCAGCCGCAGGCGATGAGCGCGCAGGCCCTGGCCGAGTCCGAGCGGGTCGCCCGGGCAGTGACCGAGGCCCTCGGCGGACGCGGGCTGTTCGGCGTCGAACTGTTCGTCAAGGGCGACCAGGTGTGGTTCAGCGAGGTGTCGCCGCGTCCGCACGATACCGGCCTGGTGACCCTGATTTCCCAGGATCTTTCCGAGTTCGCGCTGCACGCGCGGGCGATCCTCGGCCTGCCGATCCCGGTGATCCGCCAGCTCGGCCCGTCGGCCTCGGCGGTGATCCTGGTGGAAGGCAAGTCCCGGCAGGTCGCTTTCGCCAACCTCGGCGCCGCGCTGAGCGAGGCGGATACGGCGCTACGCCTGTTCGGCAAGCCGGAAGTGGATGGCCAGCGGCGGATGGGCGTGGCCCTGGCGCGCGACGAGTCGATCGACGCCGCCCGCGCCAAGGCGACCCGGGCCGCCCAGGCGGTTCGCGTCGAACTCTGA |
| Protein Properties |
| Protein Residues: |
393 |
| Protein Molecular Weight: |
42.3 kDa |
| Protein Theoretical pI: |
6.07 |
| Hydropathicity (GRAVY score): |
-0.048 |
| Charge at pH 7 (predicted): |
-4.94 |
| Protein Sequence: |
>PA3751
MTRIGTPLSPSATRVLLCGSGELGKEVAIELQRLGCEVIAVDRYGNAPAMQVAHRSHVISMLDGAALRAVIEQEKPHYIVPEIEAIATATLVELEAEGYTVVPTARAAQLTMNREGIRRLAAEELGLPTSPYHFADTFEDYRRGVERVGYPCVVKPIMSSSGKGQSVLKGPDDLQAAWDYAQEGGRAGKGRVIVEGFIDFDYEITLLTVRHVDGTTFCAPIGHRQVKGDYHESWQPQAMSAQALAESERVARAVTEALGGRGLFGVELFVKGDQVWFSEVSPRPHDTGLVTLISQDLSEFALHARAILGLPIPVIRQLGPSASAVILVEGKSRQVAFANLGAALSEADTALRLFGKPEVDGQRRMGVALARDESIDAARAKATRAAQAVRVEL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3751 and its homologs
|