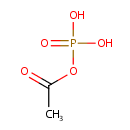
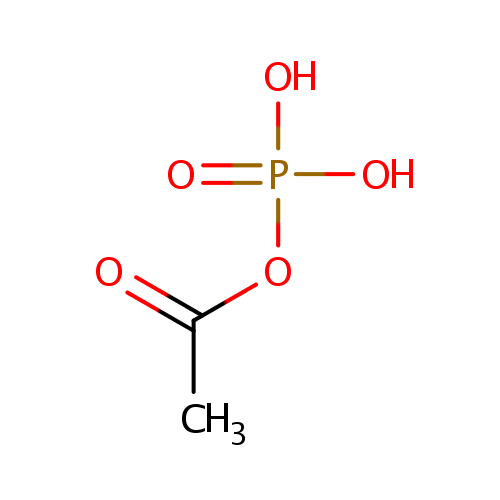
| References: |
- Bennett, B. D., Kimball, E. H., Gao, M., Osterhout, R., Van Dien, S. J., Rabinowitz, J. D. (2009). "Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli." Nat Chem Biol 5:593-599. Pubmed: 19561621
- Dowling TC, Frye RF, Zemaitis MA: Simultaneous determination of p-aminohippuric acid, acetyl-p-aminohippuric acid and iothalamate in human plasma and urine by high-performance liquid chromatography. J Chromatogr B Biomed Sci Appl. 1998 Sep 25;716(1-2):305-13. Pubmed: 9824245
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Libeer JC, Scharpe SL, Verkerk RM, Deprettere AJ, Schepens PJ: Simultaneous determination of p-aminobenzoic acid, acetyl-p-aminobenzoic acid and p-aminohippuric acid in serum and urine by capillary gas chromatography with use of a nitrogen-phosphorus detector. Clin Chim Acta. 1981 Sep 10;115(2):119-23. Pubmed: 6974621
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|