| Identification |
| Name: |
Mutator mutT protein |
| Synonyms: |
- 7,8-dihydro-8-oxoguanine-triphosphatase
- 8-oxo-dGTPase
- dGTP pyrophosphohydrolase
|
| Gene Name: |
mutT |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Replication, recombination and repair |
| Specific Function: |
Involved in the GO system responsible for removing an oxidatively damaged form of guanine (7,8-dihydro-8-oxoguanine) from DNA and the nucleotide pool. 8-oxo-dGTP is inserted opposite dA and dC residues of template DNA with almost equal efficiency thus leading to A.T to G.C transversions. MutT specifically degrades 8-oxo-dGTP to the monophosphate |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
Not Available |
| KEGG Reactions: |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
| | | | |
8-oxo-dGTP | + |  | → |  | + | 8-oxo-dGMP | + | 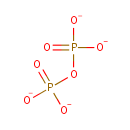 |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity | | catalytic activity | | hydrolase activity | | hydrolase activity, acting on acid anhydrides | | hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides | | nucleoside-triphosphate diphosphatase activity | | pyrophosphatase activity | | Process |
|---|
| cellular macromolecule metabolic process | | DNA metabolic process | | DNA repair | | macromolecule metabolic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4400 |
| Strand: |
- |
| Entrez Gene ID: |
881318 |
| Accession: |
NP_253090.1 |
| GI: |
15599596 |
| Sequence start: |
4930748 |
| Sequence End: |
4931695 |
| Sequence Length: |
947 |
| Gene Sequence: |
>PA4400
GTGAAACGAGTACATGTCGCCGCGGCCGTGATTCGTGGCTCCGATGGCCGGGTGCTGATCGCCCGGCGGCCGGAAGACAAGCACCAGGGCGGCCTGTGGGAGTTTCCCGGGGGCAAGGTGGAGGACGGCGAGCCGGTGCGCGCGGCGTTGGCCCGTGAGCTGGAAGAGGAACTGGGCATCCGTGTCGAGCGGGCCCGGCCGCTGATCCAGGTCCGGCACGACTATGCCGACAAGCACGTCCTGCTCGATGTCTGGGAGGTCGACGGGTTTTCCGGCGAAGCCCACGGCGCCGAGGGCCAACCGCTGGCCTGGGTGGAGCCGCGCGAGCTGGCCGACTACGAGTTCCCCGCCGCCAACGCGCCGATCGTCCAGGCCGCGCGCCTGCCGGCGCACTACCTGATCACCCCGGACGGATTGGAGCCGGGCGAGCTGATCAGCGGCGTACGCAAGGCGGTGGAGGCGGGCATCCGCCTGATCCAGTTGCGCGCGCCGAACATGTTCAGTCCCGAATACCGCGATCTCGCCATCGATATCCAGGGACTCTGCGCCGGCAAGGCGCAACTGATGCTGAAAGGCCCGCTGGAGTGGCTGGGCGACTTCCCGGCTGCCGGTTGGCACCTGACCTCCGCTCAACTGCGCAAATACGCCAGCGCCGGTCGGCCGTTCCCCGAAGGGCGCCTGCTGGCCGCCTCCTGCCACGACGCGGAGGAACTGGCCCTGGCTGCCTCGATGGGAGTGGAGTTCGTCACCCTTTCGCCGGTACAGCCGACCGAGAGCCATCCCGGCGAGCCGGCGCTGGGTTGGGACAAGGCCGCCGAACTGATCGCCGGCTTCAACCAGCCGGTCTACCTGCTGGGTGGCCTCGGTCCGCAGCAAGCCGAGCAGGCTTGGGAGCATGGAGCCCAGGGCGTGGCGGGTATCCGTGCGTTCTGGCCGGGCGGCCTTTGA |
| Protein Properties |
| Protein Residues: |
315 |
| Protein Molecular Weight: |
34 kDa |
| Protein Theoretical pI: |
5.11 |
| Hydropathicity (GRAVY score): |
-0.169 |
| Charge at pH 7 (predicted): |
-10.64 |
| Protein Sequence: |
>PA4400
MKRVHVAAAVIRGSDGRVLIARRPEDKHQGGLWEFPGGKVEDGEPVRAALARELEEELGIRVERARPLIQVRHDYADKHVLLDVWEVDGFSGEAHGAEGQPLAWVEPRELADYEFPAANAPIVQAARLPAHYLITPDGLEPGELISGVRKAVEAGIRLIQLRAPNMFSPEYRDLAIDIQGLCAGKAQLMLKGPLEWLGDFPAAGWHLTSAQLRKYASAGRPFPEGRLLAASCHDAEELALAASMGVEFVTLSPVQPTESHPGEPALGWDKAAELIAGFNQPVYLLGGLGPQQAEQAWEHGAQGVAGIRAFWPGGL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4400 and its homologs
|