| Identification |
| Name: |
Nicotinate phosphoribosyltransferase |
| Synonyms: |
|
| Gene Name: |
pncB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in nicotinate phosphoribosyltransferase activity |
| Specific Function: |
Nicotinate D-ribonucleotide + diphosphate = nicotinate + 5-phospho-alpha-D-ribose 1-diphosphate |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
- Nicotinate and nicotinamide metabolism pae00760
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
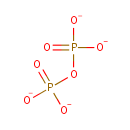
 | + |  | + |  | + | 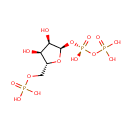 | → | 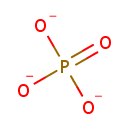 | + | Adenosine diphosphate | + | Pyrophosphate | + | nicotinate beta-D-ribonucleotide | + |  | + | 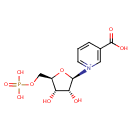 |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| catalytic activity | | nicotinate phosphoribosyltransferase activity | | transferase activity | | transferase activity, transferring glycosyl groups | | transferase activity, transferring pentosyl groups | | Process |
|---|
| cellular metabolic process | | cellular nitrogen compound metabolic process | | coenzyme biosynthetic process | | coenzyme metabolic process | | cofactor metabolic process | | metabolic process | | NAD biosynthetic process | | nicotinamide nucleotide biosynthetic process | | nicotinate nucleotide biosynthetic process | | nitrogen compound metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleoside phosphate metabolic process | | nucleotide biosynthetic process | | nucleotide metabolic process | | pyridine nucleotide biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA4919 |
| Strand: |
+ |
| Entrez Gene ID: |
882212 |
| Accession: |
NP_253606.1 |
| GI: |
15600112 |
| Sequence start: |
5518483 |
| Sequence End: |
5519682 |
| Sequence Length: |
1199 |
| Gene Sequence: |
>PA4919
ATGGCCGAAAGCGTCTTTGCCGAGCGCATTGTGCAGAACCTGCTCGATACCGATTTCTACAAGCTGACCATGATGCAGGCGGTGCTGCACAACTACCCCAACGCCGAAGTGGAGTGGGAATTCCGCTGCCGCAACGCCGAGGACCTGCGTCCGTACCTGGCGGAGATCCGCTACCAGATCGAGCGCCTCGCCGAAGTCGAGGTAACCGCCGACCAGCTCGCCTTCCTCGAACGCATCCCGTTCATGAAGCCGGACTTCATCCGCTTCCTCAGCCTGTTCCGCTTCAACCTGCGCTACGTGCACACCGGCATCGAGGATGGCCAACTGGCAATCCGCCTGCGCGGCCCATGGCTGCACGTGATCCTCTTCGAGGTGCCGCTGCTGGCCATCGTCAGCGAGGTGCGCAACCGTTATCGCTACCGCGAGGTGGTCCTGGAACAGGTCGGCGAGCAGCTCTATCGCAAGCTCGACTGGCTCTCCGCCCAGGCCAGCAGCGAGGAACTCGCGGAATTCCAGGTCGCCGACTTCGGCACCCGCCGGCGCTTCTCCTACCGCACCCAGGAGGAAGTGGTGCATATCCTCAAGCGCGACTTCCCCGGGCGCTTCGTCGGCACCAGCAACGTGCACCTGGCCCGCGAGTACGACCTCAAGCCGATCGGCACCATGGCCCACGAATGGCTGATGGCCCACCAGCAACTCGGCCCGCGGCTGGTCGACAGCCAGCAAGCGGCGCTGGACTGCTGGGTCCGCGAGTACCGCGGACAGCTCGGCATAGCCCTGACCGACTGCATTACCATGGATGCCTTCCTCGACGACTTCGACCTGTACTTCGCCAAGCTCTTCGACGGCCTGCGCCACGATTCGGGCGACCCGCTGGCCTGGGCGGAGAAAGCCATCGCCCACTACCGCCGCCTGGGTATCGACCCGCTGAGCAAGACCCTGGTGTTCTCCGACGGGCTGGACATGCCCAAGGCGTTGCAGCTGTTCCGCGCGCTACGTGGCAAGATCAACGTCAGCTTCGGCATCGGCACCAACCTGACCTGCGACATCCCGGGGGTCGAGCCGATGAACATCGTCCTGAAGATGACGGCCTGCAACGGCCATCCGGTGGCGAAGATATCCGACGCCCCGGGCAAGACCCAATGCCGCGACGAGAACTTCGTCGCCTACCTCCGCCACGTCTTCAACGTGCCGGCCTAA |
| Protein Properties |
| Protein Residues: |
399 |
| Protein Molecular Weight: |
46.1 kDa |
| Protein Theoretical pI: |
6.3 |
| Hydropathicity (GRAVY score): |
-0.153 |
| Charge at pH 7 (predicted): |
-4.52 |
| Protein Sequence: |
>PA4919
MAESVFAERIVQNLLDTDFYKLTMMQAVLHNYPNAEVEWEFRCRNAEDLRPYLAEIRYQIERLAEVEVTADQLAFLERIPFMKPDFIRFLSLFRFNLRYVHTGIEDGQLAIRLRGPWLHVILFEVPLLAIVSEVRNRYRYREVVLEQVGEQLYRKLDWLSAQASSEELAEFQVADFGTRRRFSYRTQEEVVHILKRDFPGRFVGTSNVHLAREYDLKPIGTMAHEWLMAHQQLGPRLVDSQQAALDCWVREYRGQLGIALTDCITMDAFLDDFDLYFAKLFDGLRHDSGDPLAWAEKAIAHYRRLGIDPLSKTLVFSDGLDMPKALQLFRALRGKINVSFGIGTNLTCDIPGVEPMNIVLKMTACNGHPVAKISDAPGKTQCRDENFVAYLRHVFNVPA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4919 and its homologs
|