| Identification |
| Name: |
DNA polymerase IV |
| Synonyms: |
|
| Gene Name: |
dinB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in damaged DNA binding |
| Specific Function: |
Poorly processive, error-prone DNA polymerase involved in untargeted mutagenesis. Copies undamaged DNA at stalled replication forks, which arise in vivo from mismatched or misaligned primer ends. These misaligned primers can be extended by polIV. Exhibits no 3'-5' exonuclease (proofreading) activity. Overexpression of polIV results in increased frameshift mutagenesis. It is required for stationary-phase adaptive mutation, which provides the bacterium with flexibility in dealing with environmental stress, enhancing long-term survival and evolutionary fitness. May be involved in translesional synthesis, in conjunction with the beta clamp from polIII |
| Cellular Location: |
Cytoplasm (Probable) |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
Deoxynucleoside triphosphate | + | DNA | ↔ | 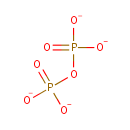 |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
Deoxynucleoside triphosphate | + | DNA(n) | → |  | + | DNA(n+1) |
| | |
Deoxynucleoside triphosphate | + | DNA | ↔ |  |
| |
|
| Complex Reactions: |
|
Deoxynucleoside triphosphate | + | DNA(n) | → |  | + | DNA(n+1) |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB000633 | Pyrophosphate | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | damaged DNA binding | | DNA binding | | DNA polymerase activity | | DNA-directed DNA polymerase activity | | nucleic acid binding | | nucleotidyltransferase activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| cellular macromolecule metabolic process | | DNA metabolic process | | DNA repair | | macromolecule metabolic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0923 |
| Strand: |
+ |
| Entrez Gene ID: |
881987 |
| Accession: |
NP_249614.1 |
| GI: |
15596120 |
| Sequence start: |
1007950 |
| Sequence End: |
1008999 |
| Sequence Length: |
1049 |
| Gene Sequence: |
>PA0923
GTGCGGAAAATCATCCATATAGACTGCGACTGTTTCTATGCCGCCCTCGAGATGCGCGACGACCCCAGCCTGCGCGGCAAGGCGCTGGCGGTTGGCGGTTCGCCGGACAAGCGCGGAGTGGTCGCCACTTGCAGCTACGAGGCGCGAGCCTATGGCGTGCGCTCGGCGATGGCCATGCGCACGGCGCTCAAGCTGTGTCCCGATCTGCTGGTGGTGCGCCCGCGTTTCGATGTCTACCGGGCGGTGTCGAAGCAGATCCATGCGATCTTCCGCGATTATACCGATCTGATCGAGCCGCTTTCGCTGGACGAGGCCTACCTGGACGTCAGCGCTTCGCCGCATTTCGCCGGTAGCGCCACGCGGATTGCCCAGGACATCCGCCGCCGGGTGGCCGAGGAGCTGCACATCACCGTTTCGGCGGGCGTCGCGCCGAACAAGTTCCTGGCCAAGATCGCCAGCGACTGGCGCAAGCCGGACGGGCTGTTCGTGATTACCCCGGAGCAGGTCGACGGGTTCGTCGCCGAGTTGCCGGTGGCCAAGTTGCATGGCGTCGGCAAGGTCACGGCGGAGCGCCTGGCGCGGATGGGCATCCGCACCTGCGCCGACCTGCGCCAGGGCAGCAAGCTGAGCCTGGTGCGCGAGTTCGGCAGCTTCGGCGAACGCCTGTGGGGACTGGCCCATGGTATCGACGAGCGACCGGTGGAAGTCGACAGCCGGCGCCAGTCGGTGAGCGTCGAATGTACCTTCGATCGCGACCTGCCGGACCTGGCGGCTTGCCTGGAGGAACTGCCGACCCTGCTCGAGGAGCTGGACGGACGCCTGCAACGCCTGGATGGCAGTTATCGGCCGGACAAGCCGTTCGTTAAACTGAAGTTTCACGATTTCACCCAGACCACGGTGGAACAGTCCGGCGCCGGGCGCGACCTGGAGAGCTACCGGCAATTGCTCGGCCAGGCCTTTGCCCGTGGCAATCGTCCGGTGCGCCTGATCGGCGTCGGCGTGCGCCTGCTCGACCTGCAGGGCGCCCACGAACAACTCAGGTTGTTCTGA |
| Protein Properties |
| Protein Residues: |
349 |
| Protein Molecular Weight: |
38.9 kDa |
| Protein Theoretical pI: |
8.07 |
| Hydropathicity (GRAVY score): |
-0.182 |
| Charge at pH 7 (predicted): |
3.72 |
| Protein Sequence: |
>PA0923
MRKIIHIDCDCFYAALEMRDDPSLRGKALAVGGSPDKRGVVATCSYEARAYGVRSAMAMRTALKLCPDLLVVRPRFDVYRAVSKQIHAIFRDYTDLIEPLSLDEAYLDVSASPHFAGSATRIAQDIRRRVAEELHITVSAGVAPNKFLAKIASDWRKPDGLFVITPEQVDGFVAELPVAKLHGVGKVTAERLARMGIRTCADLRQGSKLSLVREFGSFGERLWGLAHGIDERPVEVDSRRQSVSVECTFDRDLPDLAACLEELPTLLEELDGRLQRLDGSYRPDKPFVKLKFHDFTQTTVEQSGAGRDLESYRQLLGQAFARGNRPVRLIGVGVRLLDLQGAHEQLRLF |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0923 and its homologs
|

