| Identification |
| Name: |
Glutamate-ammonia-ligase adenylyltransferase |
| Synonyms: |
- Glutamine-synthetase adenylyltransferase
- ATase
- [Glutamate--ammonia-ligase] adenylyltransferase
|
| Gene Name: |
glnE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in [glutamate-ammonia-ligase] adenylyltransferase activity |
| Specific Function: |
Adenylation and deadenylation of glutamate--ammonia ligase |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
Not Available |
| KEGG Reactions: |
|
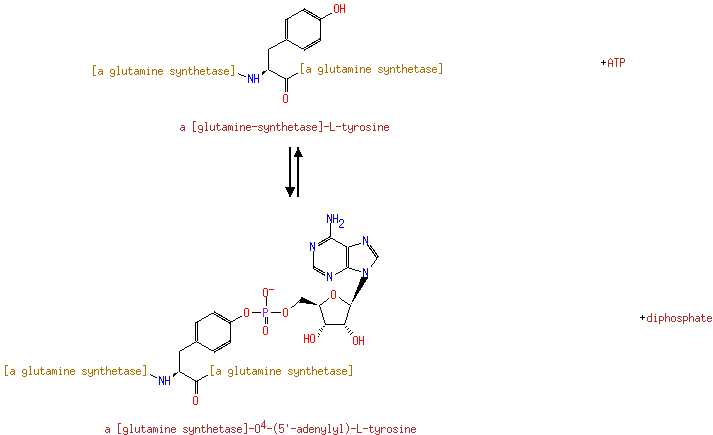
 | ↔ |  |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
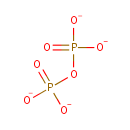
 | + | [L-glutamate:ammonia ligase (ADP-forming)] | → |  | + | adenylyl-[L-glutamate:ammonia ligase (ADP-forming)] |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenylyltransferase activity | | catalytic activity | | nucleotidyltransferase activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | [glutamate-ammonia-ligase] adenylyltransferase activity |
|
| Gene Properties |
| Locus tag: |
PA5014 |
| Strand: |
- |
| Entrez Gene ID: |
881325 |
| Accession: |
NP_253701.1 |
| GI: |
15600207 |
| Sequence start: |
5632926 |
| Sequence End: |
5635874 |
| Sequence Length: |
2948 |
| Gene Sequence: |
>PA5014
ATGAGCCTGCCTTCGCTGGCGAATCTGCCGGCCACCCTGCTGCCCGCCGCCGAACGCGCCGGGACTGCCCTCCGCTCCGCTGTCGCCGCCCTCGATGCCGCCGCCCTGGCCCGCCTGGAGGCCTGGCCCGAGGAGCGCCTGGAGGATTTCCGACGGGTCGCCGCGGCCAGCGATTTCGTCGCCGAGCAGGCAGTGCGCGACTCGGCGATGCTGCTGGAACTGGCCGAACGCGGCGAGCTGGAGAACCCCCACGCGCCCGGCGAACTGCGCAGTCAGTTGCAGGCGCGGCTGGAGGATTGCGCCGACGAGGACGAACTCGGTCGGCGCCTGCGACGTTTCCGCACACGCCAGCAACTGCGCATCATCTGGCGCGACCTGACCCGCCGCGCCGCGCTGGCGGAAACCTGTCGCGACCTGTCGGCCCTGGCCGACGCCTGCATCGACCTGGCTTGCGAGTGGCTCCATCGGCGCCAGTGCGAGCAGTTCGGCACGCCGATCGGCCGGCGCTCCGGCGAACCGCAGCGGATGGTGGTCCTCGGCATGGGCAAGCTGGGCGCGGTGGAACTCAACCTGTCGTCGGACATCGACCTGATCTTCGGTTACCCGGAGGGCGGCGAGACCGAGGGGGCGAAGCGTTCGCTGGACAACCAGGAGTTCTTCACCCGCTTGGGGCAGAAGCTGATCAAGGCGCTGGATGCGATCACCGTCGACGGCTTCGTCTTCCGCGTCGACATGCGCCTGCGTCCCTACGGCTCCAGCGGACCGCTGGTCTACAGCTTCGCCGCGCTGGAGCAGTACTACCAGGACCAGGGGCGCGACTGGGAGCGCTACGCGATGATCAAGGCGCGGGTGGTCGGCGGCGACCAGCAGGCAGGCGAGCAACTGCTCGGCATGCTGCGGCCGTTCGTCTATCGGCGCTACCTGGACTTTTCCGCCATCGAGGCGCTGCGCACCATGAAGCAGCTGATCCAGCAGGAGGTCCGGCGCAAGGGGATGTCGGAAAACATCAAGCTGGGCGAAGGCGGTATCCGCGAGGTGGAGTTCATCGCCCAGGCCTTCCAGTTGATCCATGGCGGGCGCGACCTGAGCTTGCAGCAACGGCCGCTGCTGAAGGTGCTGGCGACCCTGGAGGGGCAGGGCTACCTGCCGCCGGCGGTGGTCGAGGAGCTGCGCGGCGGCTACGAGTTCCTGCGCTACGCCGAGCATGCCATCCAGGCGCTGGCCGACCGGCAGACGCAAATGCTGCCCAGCGACGAATACGACCGCATCCGCGTGGCCTTCATCATGGGCTTCGCCAGTTGGGCCGCATTCCACGAGCGCCTGAGCCACTGGCGGGCGCGCATCGACTGGCACTTCCGCCAGGTGATCGCCGACCCGGACGAGGACGAAAGCGGCGAGGCGGCCTGCGGCAGCGTCGGCGCCGAGTGGATCCCGCTGTGGGAGGAGGCGCTGGACGAGGAGTCGGCCAGCCGGCAACTGGCCGACGCCGGCTTCGTCGATGCCGAGGCGGCCTGGAAGCGTCTCAGCGATCTGCGCCACGGCCCCCAGGTACGAGCGATGCAGCGTCTCGGCCGGGAGCGCCTGGATGCCTTCGTTCCGCGCCTGCTGGCGATGACCGTGGAAAACCCCCAGCCGGACCTGGTGCTGGAGCGGGTCCTGCCGCTGGTGGAAGCGGTGGCGCGGCGTTCCGCCTACCTGGTGTTGCTGACCGAGAACCCCGGCGCGCTGGAGCGCCTGCTGACCCTCTGCGCGGCCAGCCCGATGGTTGCCGAGCAGATCGCGCGGTTCCCGATTCTGCTCGACGAGCTGCTCAACGAGGGGCGCCTGTTCCGTCCTCCACAGGCCGCCGAGCTGGCCGCCGAGCTGCGCGAACGGCTGATGCGGATTCCCGAGGACGACCTCGAGCAGCAGATGGAGACCCTTCGTCACTTCAAGCTGGCCCACGGGCTGCGGGTGGCGGCCTCGGAGATCGCCGGCACGCTGCCGTTGATGAAGGTCAGCGACTACCTGACCTGGCTGGCCGAGGCGATCCTCGTCGAGGTCCTCGAACTGGCCTGGCGGCAGTTGGTGCAGCGTCACGGCCGGCCGTTGCGCGCCGACGGCACGCCGTGCGATCCGGACTTCGTCATCGTCGGCTACGGCAAGGTCGGCGGGCTGGAGTTCGGCCATGGCTCGGACCTCGACCTGGTGTTCATCCATGACGGCGACCCCCAGTGCGAGACCGATGGCGGCAAATCCATCGATGGCGCGCAGTTCTTTACCCGCCTGGGGCAGAAGATCATCCATTTCCTCACCGCGCAGACGCCATCCGGCACCCTCTACGAGGTCGACATGCGCCTGCGTCCGAGCGGCGCGGCCGGCCTGCTGGTGAGTTCGCTGGGCGCCTTCCAGCGCTACCAGGAGCAGGAGGCCTGGACCTGGGAGCACCAGGCGTTGGTCCGCGCCCGGGTGCTGGCCGGTTGCCGGCGGGTGCAGGCGAGCTTCGAGGCGGTACGTGCCGAGGTGCTGGCGAGGCCGCGCGATCTCGACGCGTTGCGCACCGAAGTCAGCGAAATGCGCGCCAAGATGCGCGACAACCTGGGAACCCGCGCGACCGCCGCCGGTACCGCCTCGAATGCCTTCGAGGCCACGGCAGCCTTCGATCTCAAGCACGATGCCGGTGGTATCGTCGATATCGAATTTATGGTGCAATATGCGGTTCTAGCTTGGTCCGGAGAACACCCGGCGCTGCTCGAATTCACCGATAACATCCGCATTCTGGAAGGACTGGAACGGGCTGGCCTGATCGCCAGCGAGGACGTCCGTCTCCTGCAGGAGGCCTACAAGGCCTATCGCGCAGCCGCACACCGCCTGGCACTACAGAAGGAGGCCGGGGTCGTGAGCGGCGAGCACTTCCAGACGGAGCGGCGAGAAGTGATCAGGATCTGGCGCGAGCTGCGGCTCGGCTGA |
| Protein Properties |
| Protein Residues: |
982 |
| Protein Molecular Weight: |
110.1 kDa |
| Protein Theoretical pI: |
5.22 |
| Hydropathicity (GRAVY score): |
-0.224 |
| Charge at pH 7 (predicted): |
-22.63 |
| Protein Sequence: |
>PA5014
MSLPSLANLPATLLPAAERAGTALRSAVAALDAAALARLEAWPEERLEDFRRVAAASDFVAEQAVRDSAMLLELAERGELENPHAPGELRSQLQARLEDCADEDELGRRLRRFRTRQQLRIIWRDLTRRAALAETCRDLSALADACIDLACEWLHRRQCEQFGTPIGRRSGEPQRMVVLGMGKLGAVELNLSSDIDLIFGYPEGGETEGAKRSLDNQEFFTRLGQKLIKALDAITVDGFVFRVDMRLRPYGSSGPLVYSFAALEQYYQDQGRDWERYAMIKARVVGGDQQAGEQLLGMLRPFVYRRYLDFSAIEALRTMKQLIQQEVRRKGMSENIKLGEGGIREVEFIAQAFQLIHGGRDLSLQQRPLLKVLATLEGQGYLPPAVVEELRGGYEFLRYAEHAIQALADRQTQMLPSDEYDRIRVAFIMGFASWAAFHERLSHWRARIDWHFRQVIADPDEDESGEAACGSVGAEWIPLWEEALDEESASRQLADAGFVDAEAAWKRLSDLRHGPQVRAMQRLGRERLDAFVPRLLAMTVENPQPDLVLERVLPLVEAVARRSAYLVLLTENPGALERLLTLCAASPMVAEQIARFPILLDELLNEGRLFRPPQAAELAAELRERLMRIPEDDLEQQMETLRHFKLAHGLRVAASEIAGTLPLMKVSDYLTWLAEAILVEVLELAWRQLVQRHGRPLRADGTPCDPDFVIVGYGKVGGLEFGHGSDLDLVFIHDGDPQCETDGGKSIDGAQFFTRLGQKIIHFLTAQTPSGTLYEVDMRLRPSGAAGLLVSSLGAFQRYQEQEAWTWEHQALVRARVLAGCRRVQASFEAVRAEVLARPRDLDALRTEVSEMRAKMRDNLGTRATAAGTASNAFEATAAFDLKHDAGGIVDIEFMVQYAVLAWSGEHPALLEFTDNIRILEGLERAGLIASEDVRLLQEAYKAYRAAAHRLALQKEAGVVSGEHFQTERREVIRIWRELRLG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5014 and its homologs
|