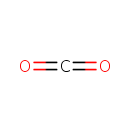
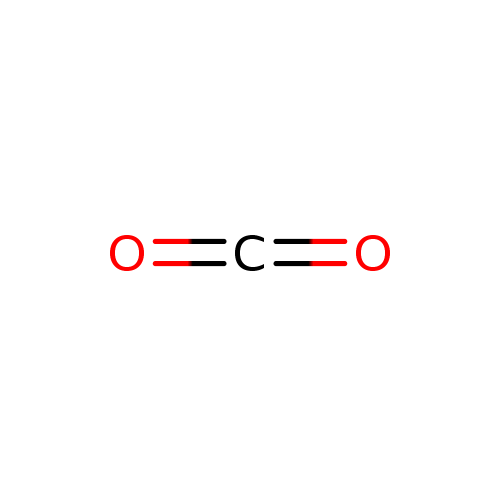Carbon dioxide (PAMDB000568)
Enzymes
- General function:
- Involved in cyanate hydratase activity
- Specific function:
- Catalyzes the reaction of cyanate with bicarbonate to produce ammonia and carbon dioxide
- Gene Name:
- cynS
- Locus Tag:
- PA2052
- Molecular weight:
- 16.8 kDa
Reactions
| Cyanate + HCO(3)(-) + 2 H(+) = NH(3) + 2 CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Meso-2,6-diaminoheptanedioate = L-lysine + CO(2)
- Gene Name:
- lysA
- Locus Tag:
- PA5277
- Molecular weight:
- 45.5 kDa
Reactions
| Meso-2,6-diaminoheptanedioate = L-lysine + CO(2). |
- General function:
- Involved in phosphoenolpyruvate carboxylase activity
- Specific function:
- Through the carboxylation of phosphoenolpyruvate (PEP) it forms oxaloacetate, a four-carbon dicarboxylic acid source for the tricarboxylic acid cycle
- Gene Name:
- ppc
- Locus Tag:
- PA3687
- Molecular weight:
- 97.8 kDa
Reactions
| Phosphate + oxaloacetate = H(2)O + phosphoenolpyruvate + HCO(3)(-). |
- General function:
- Involved in magnesium ion binding
- Specific function:
- 2 pyruvate = 2-acetolactate + CO(2)
- Gene Name:
- ilvI
- Locus Tag:
- PA4696
- Molecular weight:
- 63 kDa
Reactions
| 2 pyruvate = 2-acetolactate + CO(2). |
- General function:
- Involved in acetolactate synthase activity
- Specific function:
- 2 pyruvate = 2-acetolactate + CO(2)
- Gene Name:
- ilvH
- Locus Tag:
- PA4695
- Molecular weight:
- 17.8 kDa
Reactions
| 2 pyruvate = 2-acetolactate + CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Bifunctional enzyme that catalyzes two sequential steps of tryptophan biosynthetic pathway. The first reaction is catalyzed by the isomerase, coded by the trpF domain; the second reaction is catalyzed by the synthase, coded by the trpC domain
- Gene Name:
- trpC
- Locus Tag:
- PA0651
- Molecular weight:
- 30.3 kDa
Reactions
| N-(5-phospho-beta-D-ribosyl)anthranilate = 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate. |
| 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate = 1-C-(3-indolyl)-glycerol 3-phosphate + CO(2) + H(2)O. |
- General function:
- Involved in transferase activity, transferring acyl groups
- Specific function:
- The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2). It contains multiple copies of three enzymatic components:pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3)
- Gene Name:
- aceF
- Locus Tag:
- PA5016
- Molecular weight:
- 56.7 kDa
Reactions
| Acetyl-CoA + enzyme N(6)-(dihydrolipoyl)lysine = CoA + enzyme N(6)-(S-acetyldihydrolipoyl)lysine. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Pyruvate + ferricytochrome b1 + H(2)O = acetate + CO(2) + ferrocytochrome b1
- Gene Name:
- poxB
- Locus Tag:
- PA5297
- Molecular weight:
- 62.3 kDa
Reactions
| Pyruvate + ubiquinone + H(2)O = acetate + CO(2) + ubiquinol. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Isocitrate + NADP(+) = 2-oxoglutarate + CO(2) + NADPH
- Gene Name:
- icd
- Locus Tag:
- PA2623
- Molecular weight:
- 45.6 kDa
Reactions
| Isocitrate + NADP(+) = 2-oxoglutarate + CO(2) + NADPH. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the decarboxylation of orotidine 5'- monophosphate (OMP) to uridine 5'-monophosphate (UMP)
- Gene Name:
- pyrF
- Locus Tag:
- PA2876
- Molecular weight:
- 24.4 kDa
Reactions
| Orotidine 5'-phosphate = UMP + CO(2). |
- General function:
- Involved in metabolic process
- Specific function:
- Catalyzes the removal of elemental sulfur and selenium atoms from cysteine and selenocysteine to produce alanine. Functions as a sulfur delivery protein for NAD, biotin and Fe-S cluster synthesis. Transfers sulfur on 'Cys-456' of thiI in a transpersulfidation reaction. Transfers sulfur on 'Cys-19' of tusA in a transpersulfidation reaction. Functions also as a selenium delivery protein in the pathway for the biosynthesis of selenophosphate
- Gene Name:
- iscS
- Locus Tag:
- PA3814
- Molecular weight:
- 44.7 kDa
Reactions
| L-cysteine + acceptor = L-alanine + S-sulfanyl-acceptor. |
- General function:
- Involved in aspartate 1-decarboxylase activity
- Specific function:
- Catalyzes the pyruvoyl-dependent decarboxylation of aspartate to produce beta-alanine
- Gene Name:
- panD
- Locus Tag:
- PA4731
- Molecular weight:
- 13.9 kDa
Reactions
| L-aspartate = beta-alanine + CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the complicated ring closure reaction between the two acyclic compounds 1-deoxy-D-xylulose-5-phosphate (DXP) and 3-amino-2-oxopropyl phosphate (1-amino-acetone-3-phosphate or AAP) to form pyridoxine 5'-phosphate (PNP) and inorganic phosphate
- Gene Name:
- pdxJ
- Locus Tag:
- PA0773
- Molecular weight:
- 27.2 kDa
Reactions
| 1-deoxy-D-xylulose 5-phosphate + 3-amino-2-oxopropyl phosphate = pyridoxine 5'-phosphate + phosphate + 2 H(2)O. |
- General function:
- Involved in adenosylmethionine decarboxylase activity
- Specific function:
- Catalyzes the decarboxylation of S-adenosylmethionine to S-adenosylmethioninamine (dcAdoMet), the propylamine donor required for the synthesis of the polyamines spermine and spermidine from the diamine putrescine
- Gene Name:
- speD
- Locus Tag:
- PA0654
- Molecular weight:
- 30.4 kDa
Reactions
| S-adenosyl-L-methionine = (5-deoxy-5-adenosyl)(3-aminopropyl)-methylsulfonium salt + CO(2). |
- General function:
- Involved in phosphatidylserine decarboxylase activity
- Specific function:
- Phosphatidyl-L-serine = phosphatidylethanolamine + CO(2)
- Gene Name:
- psd
- Locus Tag:
- PA4957
- Molecular weight:
- 32 kDa
Reactions
| Phosphatidyl-L-serine = phosphatidylethanolamine + CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the condensation reaction of fatty acid synthesis by the addition to an acyl acceptor of two carbons from malonyl-ACP. Specific for elongation from C-10 to unsaturated C-16 and C-18 fatty acids
- Gene Name:
- fabB
- Locus Tag:
- PA1609
- Molecular weight:
- 42.8 kDa
Reactions
| Acyl-[acyl-carrier-protein] + malonyl-[acyl-carrier-protein] = 3-oxoacyl-[acyl-carrier-protein] + CO(2) + [acyl-carrier-protein]. |
- General function:
- Involved in chorismate mutase activity
- Specific function:
- Chorismate = prephenate
- Gene Name:
- pheA
- Locus Tag:
- PA3166
- Molecular weight:
- 40.6 kDa
Reactions
| Chorismate = prephenate. |
| Prephenate = phenylpyruvate + H(2)O + CO(2). |
- General function:
- Involved in transferase activity, transferring acyl groups other than amino-acyl groups
- Specific function:
- Catalyzes the condensation reaction of fatty acid synthesis by the addition to an acyl acceptor of two carbons from malonyl-ACP. Has a preference for short chain acid substrates and may function to supply the octanoic substrates for lipoic acid biosynthesis
- Gene Name:
- fabF
- Locus Tag:
- PA2965
- Molecular weight:
- 43.5 kDa
Reactions
| (Z)-hexadec-11-enoyl-[acyl-carrier-protein] + malonyl-[acyl-carrier-protein] = (Z)-3-oxooctadec-13-enoyl-[acyl-carrier-protein] + CO(2) + [acyl-carrier-protein]. |
- General function:
- Involved in electron carrier activity
- Specific function:
- Formate dehydrogenase allows Pseudomonas aeruginosa to use formate as major electron donor during anaerobic respiration, when nitrate is used as electron acceptor. The beta chain is an electron transfer unit containing 4 cysteine clusters involved in the formation of iron-sulfur centers. Electrons are transferred from the gamma chain to the molybdenum cofactor of the alpha subunit
- Gene Name:
- fdnH
- Locus Tag:
- PA4811
- Molecular weight:
- 33.8 kDa
- General function:
- Involved in carbonate dehydratase activity
- Specific function:
- Reversible hydration of carbon dioxide. Carbon dioxide formed in the bicarbonate-dependent decomposition of cyanate by cyanase (cynS) diffuses out of the cell faster than it would be hydrated to bicarbonate, so the apparent function of this enzyme is to catalyze the hydration of carbon dioxide and thus prevent depletion of cellular bicarbonate
- Gene Name:
- cynT
- Locus Tag:
- PA2053
- Molecular weight:
- 23.4 kDa
Reactions
| H(2)CO(3) = CO(2) + H(2)O. |
- General function:
- Involved in transferase activity, transferring phosphorus-containing groups
- Specific function:
- CTP + phosphatidate = diphosphate + CDP- diacylglycerol
- Gene Name:
- cdsA
- Locus Tag:
- PA3651
- Molecular weight:
- 28.9 kDa
Reactions
| CTP + phosphatidate = diphosphate + CDP-diacylglycerol. |
- General function:
- Involved in phosphopantothenate--cysteine ligase activity
- Specific function:
- Catalyzes two steps in the biosynthesis of coenzyme A. In the first step cysteine is conjugated to 4'-phosphopantothenate to form 4-phosphopantothenoylcysteine, in the latter compound is decarboxylated to form 4'-phosphopantotheine
- Gene Name:
- coaBC
- Locus Tag:
- PA5320
- Molecular weight:
- 43.1 kDa
Reactions
| N-((R)-4'-phosphopantothenoyl)-L-cysteine = pantotheine 4'-phosphate + CO(2). |
| CTP + (R)-4'-phosphopantothenate + L-cysteine = CMP + diphosphate + N-((R)-4'-phosphopantothenoyl)-L-cysteine. |
- General function:
- Involved in respiratory electron transport chain
- Specific function:
- Formate dehydrogenase allows Pseudomonas aeruginosa to use formate as major electron donor during anaerobic respiration, when nitrate is used as electron acceptor. Subunit gamma is the cytochrome b556(FDN) component of the formate dehydrogenase
- Gene Name:
- fdnI
- Locus Tag:
- PA4810
- Molecular weight:
- 23.9 kDa
- General function:
- Involved in tartronate-semialdehyde synthase activity
- Specific function:
- Catalyzes the condensation of two molecules of glyoxylate to give 2-hydroxy-3-oxopropanoate (also termed tartronate semialdehyde)
- Gene Name:
- gcl
- Locus Tag:
- PA1502
- Molecular weight:
- 64.4 kDa
Reactions
| 2 glyoxylate = tartronate semialdehyde + CO(2). |
- General function:
- Involved in oxoglutarate dehydrogenase (succinyl-transferring) activity
- Specific function:
- The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components:2- oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3)
- Gene Name:
- sucA
- Locus Tag:
- PA1585
- Molecular weight:
- 105.9 kDa
Reactions
| 2-oxoglutarate + [dihydrolipoyllysine-residue succinyltransferase] lipoyllysine = [dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine + CO(2). |
- General function:
- Involved in transferase activity, transferring acyl groups
- Specific function:
- The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components:2- oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3)
- Gene Name:
- sucB
- Locus Tag:
- PA1586
- Molecular weight:
- 42.9 kDa
Reactions
| Succinyl-CoA + enzyme N(6)-(dihydrolipoyl)lysine = CoA + enzyme N(6)-(S-succinyldihydrolipoyl)lysine. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2). It contains multiple copies of three enzymatic components:pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3)
- Gene Name:
- aceE
- Locus Tag:
- PA5015
- Molecular weight:
- 99.6 kDa
Reactions
| Pyruvate + [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine = [dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine + CO(2). |
- General function:
- Involved in 8-amino-7-oxononanoate synthase activity
- Specific function:
- Catalyzes the decarboxylative condensation of pimeloyl- CoA and L-alanine to produce 8-amino-7-oxononanoate (AON), coenzyme A, and carbon dioxide
- Gene Name:
- bioF
- Locus Tag:
- PA0501
- Molecular weight:
- 42.2 kDa
Reactions
| 6-carboxyhexanoyl-CoA + L-alanine = 8-amino-7-oxononanoate + CoA + CO(2). |
- General function:
- Involved in magnesium ion binding
- Specific function:
- ATP + 7,8-diaminononanoate + CO(2) = ADP + phosphate + dethiobiotin
- Gene Name:
- bioD
- Locus Tag:
- PA0504
- Molecular weight:
- 23.3 kDa
Reactions
| ATP + 7,8-diaminononanoate + CO(2) = ADP + phosphate + dethiobiotin. |
- General function:
- Involved in phosphatidylglycerophosphatase activity
- Specific function:
- One of the three phospholipid phosphatases, specifically hydrolyzes phosphatidylglycerophosphate
- Gene Name:
- pgpA
- Locus Tag:
- PA4050
- Molecular weight:
- 19.6 kDa
Reactions
| Phosphatidylglycerophosphate + H(2)O = phosphatidylglycerol + phosphate. |
- General function:
- Involved in 4-hydroxythreonine-4-phosphate dehydrogenase activity
- Specific function:
- Catalyzes the NAD(P)-dependent oxidation of 4- (phosphohydroxy)-L-threonine (HTP) into 2-amino-3-oxo-4- (phosphohydroxy)butyric acid which spontaneously decarboxylates to form 3-amino-2-oxopropyl phosphate (AHAP)
- Gene Name:
- pdxA
- Locus Tag:
- PA0593
- Molecular weight:
- 34.9 kDa
Reactions
| 4-(phosphonooxy)-L-threonine + NAD(+) = (2S)-2-amino-3-oxo-4-phosphonooxybutanoate + NADH. |
- General function:
- Involved in carboxy-lyase activity
- Specific function:
- L-ornithine = putrescine + CO(2)
- Gene Name:
- speC
- Locus Tag:
- PA4519
- Molecular weight:
- 43.6 kDa
Reactions
| L-ornithine = putrescine + CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the biosynthesis of agmatine from arginine
- Gene Name:
- speA
- Locus Tag:
- PA4839
- Molecular weight:
- 70.7 kDa
Reactions
| L-arginine = agmatine + CO(2). |
- General function:
- Involved in phosphoenolpyruvate carboxykinase (ATP) activity
- Specific function:
- ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO(2)
- Gene Name:
- pckA
- Locus Tag:
- PA5192
- Molecular weight:
- 55.7 kDa
Reactions
| ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO(2). |
- General function:
- Involved in ligase activity
- Specific function:
- This protein is a component of the acetyl coenzyme A carboxylase complex; first, biotin carboxylase catalyzes the carboxylation of the carrier protein and then the transcarboxylase transfers the carboxyl group to form malonyl-CoA
- Gene Name:
- accC
- Locus Tag:
- PA4848
- Molecular weight:
- 48.9 kDa
Reactions
| ATP + biotin-[carboxyl-carrier-protein] + CO(2) = ADP + phosphate + carboxy-biotin-[carboxyl-carrier-protein]. |
| ATP + acetyl-CoA + HCO(3)(-) = ADP + phosphate + malonyl-CoA. |
- General function:
- Involved in formate dehydrogenase (NAD+) activity
- Specific function:
- Formate dehydrogenase allows Pseudomonas aeruginosa to use formate as major electron donor during anaerobic respiration, when nitrate is used as electron acceptor. The alpha subunit forms the active site
- Gene Name:
- fdnG
- Locus Tag:
- PA4812
- Molecular weight:
- 104.7 kDa
Reactions
| Formate + NAD(+) = CO(2) + NADH. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- (S)-malate + NAD(+) = pyruvate + CO(2) + NADH
- Gene Name:
- sfcA
- Locus Tag:
- PA3471
- Molecular weight:
- 62.4 kDa
Reactions
| (S)-malate + NAD(+) = pyruvate + CO(2) + NADH. |
- General function:
- Involved in aminomethyltransferase activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine
- Gene Name:
- gcvT
- Locus Tag:
- PA5215
- Molecular weight:
- 38.9 kDa
Reactions
| [Protein]-S(8)-aminomethyldihydrolipoyllysine + tetrahydrofolate = [protein]-dihydrolipoyllysine + 5,10-methylenetetrahydrofolate + NH(3). |
- General function:
- Involved in uroporphyrinogen decarboxylase activity
- Specific function:
- Catalyzes the decarboxylation of four acetate groups of uroporphyrinogen-III to yield coproporphyrinogen-III
- Gene Name:
- hemE
- Locus Tag:
- PA5034
- Molecular weight:
- 38.8 kDa
Reactions
| Uroporphyrinogen III = coproporphyrinogen + 4 CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Involved in the catabolism of quinolinic acid (QA)
- Gene Name:
- nadC
- Locus Tag:
- PA4524
- Molecular weight:
- 30.6 kDa
Reactions
| Beta-nicotinate D-ribonucleotide + diphosphate + CO(2) = pyridine-2,3-dicarboxylate + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the oxidation of 3-carboxy-2-hydroxy-4- methylpentanoate (3-isopropylmalate) to 3-carboxy-4-methyl-2- oxopentanoate. The product decarboxylates to 4-methyl-2 oxopentanoate
- Gene Name:
- leuB
- Locus Tag:
- PA3118
- Molecular weight:
- 39.1 kDa
Reactions
| (2R,3S)-3-isopropylmalate + NAD(+) = 4-methyl-2-oxopentanoate + CO(2) + NADH. |
- General function:
- Involved in catalytic activity
- Specific function:
- Condenses 4-methyl-5-(beta-hydroxyethyl)thiazole monophosphate (THZ-P) and 2-methyl-4-amino-5-hydroxymethyl pyrimidine pyrophosphate (HMP-PP) to form thiamine monophosphate (TMP)
- Gene Name:
- thiE
- Locus Tag:
- PA3976
- Molecular weight:
- 22.1 kDa
Reactions
| 2-methyl-4-amino-5-hydroxymethylpyrimidine diphosphate + 4-methyl-5-(2-phosphono-oxyethyl)thiazole = diphosphate + thiamine phosphate. |
- General function:
- Involved in coproporphyrinogen oxidase activity
- Specific function:
- Anaerobic transformation of coproporphyrinogen-III into protoporphyrinogen-IX
- Gene Name:
- hemN
- Locus Tag:
- PA1546
- Molecular weight:
- 52.5 kDa
Reactions
| Coproporphyrinogen-III + 2 S-adenosyl-L-methionine = protoporphyrinogen-IX + 2 CO(2) + 2 L-methionine + 2 5'-deoxyadenosine. |
- General function:
- Involved in glycine dehydrogenase (decarboxylating) activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The P protein binds the alpha-amino group of glycine through its pyridoxal phosphate cofactor; CO(2) is released and the remaining methylamine moiety is then transferred to the lipoamide cofactor of the H protein
- Gene Name:
- gcvP
- Locus Tag:
- PA5213
- Molecular weight:
- 104.7 kDa
Reactions
| Glycine + H-protein-lipoyllysine = H-protein-S-aminomethyldihydrolipoyllysine + CO(2). |
- General function:
- Involved in coproporphyrinogen oxidase activity
- Specific function:
- Key enzyme in heme biosynthesis. Catalyzes the oxidative decarboxylation of propionic acid side chains of rings A and B of coproporphyrinogen III
- Gene Name:
- hemF
- Locus Tag:
- PA0024
- Molecular weight:
- 34.8 kDa
Reactions
| Coproporphyrinogen-III + O(2) + 2 H(+) = protoporphyrinogen-IX + 2 CO(2) + 2 H(2)O. |
- General function:
- Involved in cellular amino acid biosynthetic process
- Specific function:
- ATP + NH(3) + CO(2) = ADP + carbamoyl phosphate
- Gene Name:
- arcC
- Locus Tag:
- PA5173
- Molecular weight:
- 33.1 kDa
Reactions
| ATP + NH(3) + CO(2) = ADP + carbamoyl phosphate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the conversion of taurine and alpha ketoglutarate to sulfite, aminoacetaldehyde and succinate. Required for the utilization of taurine (2-aminoethanesulfonic acid) as an alternative sulfur source. Pentane-sulfonic acid, 3- (N-morpholino)propanesulfonic acid and 1,3-dioxo-2- isoindolineethanesulfonic acid are also substrates for this enzyme
- Gene Name:
- tauD
- Locus Tag:
- PA3935
- Molecular weight:
- 31 kDa
Reactions
| Taurine + 2-oxoglutarate + O(2) = sulfite + aminoacetaldehyde + succinate + CO(2). |
- General function:
- Involved in coproporphyrinogen oxidase activity
- Specific function:
- Not Available
- Gene Name:
- yggW
- Locus Tag:
- PA0386
- Molecular weight:
- 42.5 kDa
- General function:
- Involved in carbonate dehydratase activity
- Specific function:
- H(2)CO(3) = CO(2) + H(2)O
- Gene Name:
- can
- Locus Tag:
- PA4676
- Molecular weight:
- 24.2 kDa
Reactions
| H(2)CO(3) = CO(2) + H(2)O. |
- General function:
- Involved in N-succinylarginine dihydrolase activity
- Specific function:
- Catalyzes the hydrolysis of N(2)-succinylarginine into N(2)-succinylornithine, ammonia and CO(2)
- Gene Name:
- astB
- Locus Tag:
- PA0899
- Molecular weight:
- 48.9 kDa
Reactions
| N(2)-succinyl-L-arginine + 2 H(2)O = N(2)-succinyl-L-ornithine + 2 NH(3) + CO(2). |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- (S)-malate + NADP(+) = pyruvate + CO(2) + NADPH
- Gene Name:
- maeB
- Locus Tag:
- PA5046
- Molecular weight:
- 45.4 kDa
Reactions
| (S)-malate + NADP(+) = pyruvate + CO(2) + NADPH. |
- General function:
- Involved in hydroxymethyl-, formyl- and related transferase activity
- Specific function:
- Bifunctional enzyme that catalyzes the oxidative decarboxylation of UDP-glucuronic acid (UDP-GlcUA) to UDP-4-keto- arabinose (UDP-Ara4O) and the addition of a formyl group to UDP-4- amino-4-deoxy-L-arabinose (UDP-L-Ara4N) to form UDP-L-4-formamido- arabinose (UDP-L-Ara4FN). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides
- Gene Name:
- arnA
- Locus Tag:
- PA3554
- Molecular weight:
- 74.4 kDa
Reactions
| UDP-alpha-D-glucuronate + NAD(+) = UDP-beta-L-threo-pentapyranos-4-ulose + CO(2) + NADH. |
| 10-formyltetrahydrofolate + UDP-4-amino-4-deoxy-beta-L-arabinose = 5,6,7,8-tetrahydrofolate + UDP-4-deoxy-4-formamido-beta-L-arabinose. |
- General function:
- Involved in 1-deoxy-D-xylulose-5-phosphate synthase activity
- Specific function:
- Catalyzes the acyloin condensation reaction between C atoms 2 and 3 of pyruvate and glyceraldehyde 3-phosphate to yield 1-deoxy-D-xylulose-5-phosphate (DXP)
- Gene Name:
- dxs
- Locus Tag:
- PA4044
- Molecular weight:
- 68 kDa
Reactions
| Pyruvate + D-glyceraldehyde 3-phosphate = 1-deoxy-D-xylulose 5-phosphate + CO(2). |
- General function:
- Involved in ureidoglycolate hydrolase activity
- Specific function:
- Involved in the anaerobic utilization of allantoin. Reinforces the induction of genes involved in the degradation of allantoin and glyoxylate by producing glyoxylate
- Gene Name:
- allA
- Locus Tag:
- PA1514
- Molecular weight:
- 19 kDa
Reactions
| (S)-ureidoglycolate + H(2)O = glyoxylate + 2 NH(3) + CO(2). |
- General function:
- Involved in carboxy-lyase activity
- Specific function:
- Catalyzes the decarboxylation of 3-octaprenyl-4-hydroxy benzoate to 2-octaprenylphenol
- Gene Name:
- ubiD
- Locus Tag:
- PA5237
- Molecular weight:
- 54.6 kDa
- General function:
- Involved in RNA binding
- Specific function:
- Catalyzes the ATP-dependent transfer of a sulfur to tRNA to produce 4-thiouridine in position 8 of tRNAs, which functions as a near-UV photosensor. Also catalyzes the transfer of sulfur to the sulfur carrier protein ThiS, forming ThiS-thiocarboxylate. This is a step in the synthesis of thiazole, in the thiamine biosynthesis pathway. The sulfur is donated as persulfide by iscS
- Gene Name:
- thiI
- Locus Tag:
- PA5118
- Molecular weight:
- 54.8 kDa
Reactions
| L-cysteine + 'activated' tRNA = L-serine + tRNA containing a thionucleotide. |
| [IscS]-SSH + [ThiS]-COAMP = [IscS]-SH + [ThiS]-COSH + AMP. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the NADP-dependent oxidation of L-allo- threonine to L-2-amino-3-keto-butyrate, which is spontaneously decarboxylated into aminoacetone. Also acts on L-serine, D-serine, D-threonine, D-3-hydroxyisobutyrate, L-3-hydroxyisobutyrate, D- glycerate and L-glycerate
- Gene Name:
- ydfG
- Locus Tag:
- PA4907
- Molecular weight:
- 27.4 kDa
Reactions
| 3-hydroxypropanoate + NADP(+) = 3-oxopropanoate + NADPH. |
- General function:
- Involved in phosphotransferase activity, for other substituted phosphate groups
- Specific function:
- CDP-diacylglycerol + L-serine = CMP + (3-sn- phosphatidyl)-L-serine
- Gene Name:
- pssA
- Locus Tag:
- PA4693
- Molecular weight:
- 29.3 kDa
Reactions
| CDP-diacylglycerol + L-serine = CMP + (3-sn-phosphatidyl)-L-serine. |
- General function:
- Amino acid transport and metabolism
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The H protein shuttles the methylamine group of glycine from the P protein to the T protein
- Gene Name:
- gcvH
- Locus Tag:
- PA5214
- Molecular weight:
- 13.6 kDa