| Identification |
| Name: |
Pyruvate dehydrogenase E1 component |
| Synonyms: |
Not Available |
| Gene Name: |
aceE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in oxidoreductase activity |
| Specific Function: |
The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2). It contains multiple copies of three enzymatic components:pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3) |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
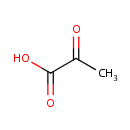 | + | Enzyme N6-(lipoyl)lysine | ↔ | [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine | + | 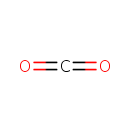 | + | [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine |
| Pyruvic acid + Enzyme N6-(lipoyl)lysine ↔ [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine + Carbon dioxide + [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine ReactionCard |
|
| SMPDB Reactions: |
|
 | + | a [pyruvate dehydrogenase E2 protein] N6-lipoyl-L-lysine | + |  | → | a [pyruvate dehydrogenase E2 protein] N6-S-acetyldihydrolipoyl-L-lysine | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
 | + | [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine | → | [dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine | + |  |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | oxidoreductase activity | | Process |
|---|
| metabolic process | | oxidation reduction |
|
| Gene Properties |
| Locus tag: |
PA5015 |
| Strand: |
+ |
| Entrez Gene ID: |
881326 |
| Accession: |
NP_253702.1 |
| GI: |
15600208 |
| Sequence start: |
5636156 |
| Sequence End: |
5638804 |
| Sequence Length: |
2648 |
| Gene Sequence: |
>PA5015
ATGCAAGACCTCGATCCCGTCGAAACCCAGGAATGGCTGGACGCCCTGGAGTCCGTACTCGACCGCGAAGGTGAAGACCGCGCTCATTACCTGATGACCCGCATGGGCGAGCTGGCCAGCCGTAGCGGCACCCAACTGCCCTACGCGATCACCACGCCCTATCGCAACACCATTCCGGTAACCCACGAAGCACGCATGCCCGGCGACCTGTTCATGGAACGCCGGATCCGCTCCCTGGTGCGCTGGAACGCCCTGGCGATGGTGATGCGGGCGAACAAGCACGACCCGGACCTGGGCGGCCACATCTCCACCTTCGCCTCCTCGGCGACCCTCTACGACATCGGCTTCAACTATTTCTTCCAGGCTCCCACCGACGAACACGGCGGCGACCTGGTGTTCTTCCAGGGCCACGCGTCCCCCGGCGTCTACGCCCGGGCCTTCCTCGAAGGCCGCATCAGCGAGGAACAACTGGAGAACTTCCGCCAGGAAGTGGACGGCAACGGCCTGTCCTCCTATCCGCACCCCTGGCTGATGCCAGACTTCTGGCAGTTCCCCACCGTATCCATGGGCCTTGGCCCGATCCAGGCGATCTACCAGGCACGCTTCATGAAGTACCTGGAAAGCCGCGGCTTCATCCCCGCCGGCAAGCAGAAGGTCTGGTGCTTCATGGGCGACGGCGAGTGCGACGAGCCGGAATCCCTCGGCGCGATCTCCCTGGCCGGCCGCGAGAAACTCGACAACCTGATCTTCGTCATCAACTGCAACCTGCAGCGCCTCGACGGCCCGGTCCGCGGCAACGCCAAGATCATCCAGGAACTGGAAGGCGTGTTCCGCGGCGCCGAGTGGAACGTCAACAAGGTCATCTGGGGTCGCTTCTGGGACCCGCTGTTCGCCAAGGACACCGCCGGCCTGCTGCAGCAGCGCATGGACGAGGTCATCGACGGCGAATACCAGAACTACAAGGCGAAAGACGGCGCCTACGTGCGCGAGCACTTCTTCGGCGCGCGTCCGGAACTGCTGGAAATGGTCAAGGACCTTTCCGACGAGGAAATCTGGAAGCTCAACCGCGGCGGCCACGACCCCTACAAGGTCTATGCGGCCTACCACCAGGCGGTCAACCACAAGGGCCAGCCGACCGTCATCCTGGCCAAGACCATCAAGGGCTACGGCACCGGCAGCGGCGAAGCGAAGAACATCGCGCACAACGTGAAGAAGGTCGACGTCGACAGCCTGCGCGCGTTCCGCGACAAGTTCGACATCCCGGTCAAGGACGCCGACCTGGAGAAGCTGCCGTTCTACAAGCCTGAAGAAGGCAGCGCCGAGGCCAAGTACCTGGCCGAGCGCCGCGCCGCGCTGGGCGGCTTCATGCCGGTGCGCCGGCAGAAGAGCATGAGCGTGCCGGTTCCGCCGCTGGAAACCCTGAAGGCCATGCTCGACGGCTCCGGCGATCGCGAGATCTCCACCACCATGGCCTTCGTGCGGATCATCTCGCAGCTGGTCAAGGACAAGGAACTCGGCCCGCGCATCGTCCCGATCGTCCCGGACGAGGCCCGTACCTTCGGCATGGAAGGCATGTTCCGCCAGCTCGGCATCTACTCCTCGGTCGGCCAGCTCTACGAGCCGGTGGACAAGGACCAGGTGATGTTCTACCGCGAGGACAAGAAGGGCCAGATCCTCGAGGAAGGCATCAACGAGGCCGGCGCCATGTCCAGCTGGATTGCCGCCGGTACCTCCTACAGCACCCACAACCAGCCGATGCTGCCGTTCTACATCTTCTATTCGATGTTCGGCTTCCAGCGCATCGGCGACCTGGCCTGGGCCGCCGGCGACAGCCGCGCGCACGGCTTCCTGATCGGCGGCACCGCCGGTCGCACCACGCTGAACGGCGAAGGCCTGCAGCACGAGGACGGCCACAGCCACCTGCTGGCCTCGACCATCCCGAACTGCCGCACCTACGATCCGACCTACGCCTACGAACTGGCGGTGATCATCCGCGAAGGCTCGCGGCAGATGATCGAAGAGCAGCAGGACATCTTCTACTACATCACCGTGATGAACGAGAACTACGTCCAGCCGGCCATGCCGAAGGGCGCAGAGGAAGGCATCATCAAGGGCATGTACCTCCTCGAGGAGGACAAGAAGGAAGCCGCTCACCACGTGCAACTGCTGGGCTCCGGCACCATCCTGCGCGAAGTCGAGGAAGCGGCCAAGCTGCTGCGCAACGACTTCGGCATCGGCGCCGACGTCTGGAGCGTGCCGAGCTTCAACGAGCTGCGTCGCGACGGCCTTGCCGTCGAGCGCTGGAACCGCCTGCATCCGGGCCAGAAGCCGAAGCAGAGCTACGTCGAGGAGTGCCTGGGCGGCCGTCGCGGCCCGGTGATCGCCTCCACCGACTACATGAAGCTCTACGCCGAGCAGATCCGCCAGTGGGTTCCCTCCAAGGAGTACAAGGTGCTCGGCACCGACGGCTTCGGCCGCAGCGACAGCCGCAAGAAGCTGCGCAACTTCTTCGAAGTGGATCGTCACTGGGTGGTGCTGGCCGCGCTCGAAGCGCTGGCCGATCGCGGCGATATCGAGCCGAAAGTGGTGGCCGAGGCCATCGCCAAGTACGGTATCGATCCGGAGAAGCGCAACCCGCTGGATTGCTAA |
| Protein Properties |
| Protein Residues: |
882 |
| Protein Molecular Weight: |
99.6 kDa |
| Protein Theoretical pI: |
5.59 |
| Hydropathicity (GRAVY score): |
-0.426 |
| Charge at pH 7 (predicted): |
-17.07 |
| Protein Sequence: |
>PA5015
MQDLDPVETQEWLDALESVLDREGEDRAHYLMTRMGELASRSGTQLPYAITTPYRNTIPVTHEARMPGDLFMERRIRSLVRWNALAMVMRANKHDPDLGGHISTFASSATLYDIGFNYFFQAPTDEHGGDLVFFQGHASPGVYARAFLEGRISEEQLENFRQEVDGNGLSSYPHPWLMPDFWQFPTVSMGLGPIQAIYQARFMKYLESRGFIPAGKQKVWCFMGDGECDEPESLGAISLAGREKLDNLIFVINCNLQRLDGPVRGNAKIIQELEGVFRGAEWNVNKVIWGRFWDPLFAKDTAGLLQQRMDEVIDGEYQNYKAKDGAYVREHFFGARPELLEMVKDLSDEEIWKLNRGGHDPYKVYAAYHQAVNHKGQPTVILAKTIKGYGTGSGEAKNIAHNVKKVDVDSLRAFRDKFDIPVKDADLEKLPFYKPEEGSAEAKYLAERRAALGGFMPVRRQKSMSVPVPPLETLKAMLDGSGDREISTTMAFVRIISQLVKDKELGPRIVPIVPDEARTFGMEGMFRQLGIYSSVGQLYEPVDKDQVMFYREDKKGQILEEGINEAGAMSSWIAAGTSYSTHNQPMLPFYIFYSMFGFQRIGDLAWAAGDSRAHGFLIGGTAGRTTLNGEGLQHEDGHSHLLASTIPNCRTYDPTYAYELAVIIREGSRQMIEEQQDIFYYITVMNENYVQPAMPKGAEEGIIKGMYLLEEDKKEAAHHVQLLGSGTILREVEEAAKLLRNDFGIGADVWSVPSFNELRRDGLAVERWNRLHPGQKPKQSYVEECLGGRRGPVIASTDYMKLYAEQIRQWVPSKEYKVLGTDGFGRSDSRKKLRNFFEVDRHWVVLAALEALADRGDIEPKVVAEAIAKYGIDPEKRNPLDC |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5015 and its homologs
|