| Identification |
| Name: |
3-isopropylmalate dehydrogenase |
| Synonyms: |
- 3-IPM-DH
- Beta-IPM dehydrogenase
- IMDH
|
| Gene Name: |
leuB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in magnesium ion binding |
| Specific Function: |
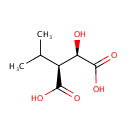
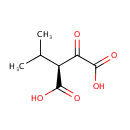
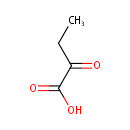
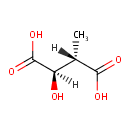
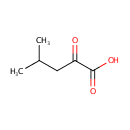
Catalyzes the oxidation of 3-carboxy-2-hydroxy-4- methylpentanoate (3-isopropylmalate) to 3-carboxy-4-methyl-2- oxopentanoate. The product decarboxylates to 4-methyl-2 oxopentanoate |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
- C5-Branched dibasic acid metabolism pae00660
- Metabolic pathways pae01100
- Valine, leucine and isoleucine biosynthesis pae00290
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| 3-isopropylmalate dehydrogenase activity | | binding | | catalytic activity | | cation binding | | ion binding | | magnesium ion binding | | metal ion binding | | NAD or NADH binding | | nucleotide binding | | oxidoreductase activity | | oxidoreductase activity, acting on CH-OH group of donors | | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | | Process |
|---|
| branched chain family amino acid metabolic process | | cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process | | cellular metabolic process | | leucine biosynthetic process | | leucine metabolic process | | metabolic process | | oxidation reduction |
|
| Gene Properties |
| Locus tag: |
PA3118 |
| Strand: |
- |
| Entrez Gene ID: |
882812 |
| Accession: |
NP_251808.1 |
| GI: |
15598314 |
| Sequence start: |
3500668 |
| Sequence End: |
3501750 |
| Sequence Length: |
1082 |
| Gene Sequence: |
>PA3118
ATGAGCAAGCAGATCCTGGTTCTCCCCGGTGACGGTATCGGCCCGGAAATCATGGCGGAAGCGGTCAAGGTGCTGGAACTGGCCAACGACAGGTTCCAGCTCGGCTTCGAACTGGCCGAGGACGTCATCGGTGGCGCGGCCATCGACAAGCACGGCGTGCCGCTGGCCGACCAGACCCTGCAGCGCGCGCGCCAGGCCGATGCGGTGCTGCTGGGCGCGGTAGGGGGGCCGAAATGGGACAGGATCGAACGCGATATCCGTCCCGAGCGCGGTCTGCTGAAGATTCGCTCGCAACTGGGCCTGTTCGCCAACCTGCGTCCGGCCATCCTCTATCCGCAACTGGCCGCGGCCTCCAGCCTGAAGCCGGAAGTGGTCGCCGGGCTGGACATCCTCATCGTCCGCGAACTGACCGGCGGCATCTACTTCGGCCAGCCCCGCGAGCAGCGCGTGCTGGAGAACGGCGAGCGCCAGGCCTACGACACCCTGCCGTACAGCGAGAGCGAAATCCGTCGTATCGCCCGCGTCGGTTTCGACATGGCCCGCGTACGCAACAACAGGCTCTGCTCGGTGGACAAGGCCAACGTTCTCGCCTCCAGCCAACTGTGGCGGGAAGTGGTGGAAGAGGTGGCGAAGGACTACCCGGACGTCGAGCTTTCCCATATGTACGTGGATAACGCCGCCATGCAACTGGTGCGCGCGCCCAAGCAGTTCGACGTGATGGTCACCGACAACATGTTCGGCGACATCCTGTCGGATGAGGCTTCCATGCTGACCGGTTCCATCGGCATGCTGCCTTCTGCCTCGCTCGACGCGAACAACAAGGGCATGTACGAACCCTGCCACGGTTCCGCCCCGGACATCGCCGGTCAGGGCATCGCCAACCCGTTGGCGACCATCCTCTCGGTCTCGATGATGTTGCGCTACAGCTTCAGCCAGGCGACCGCCGCCGACGCCATCGAGCAGGCGGTGAGCAAGGTCCTCGATCAGGGGCTGCGCACCGGCGACATCTGGTCCGAAGGCTGCCGCAAGGTCGGCACCCGGGAAATGGGCGATGCCGTAGTCGCGGCACTCAAGAATCTGTAA |
| Protein Properties |
| Protein Residues: |
360 |
| Protein Molecular Weight: |
39.1 kDa |
| Protein Theoretical pI: |
4.77 |
| Hydropathicity (GRAVY score): |
-0.067 |
| Charge at pH 7 (predicted): |
-9.36 |
| Protein Sequence: |
>PA3118
MSKQILVLPGDGIGPEIMAEAVKVLELANDRFQLGFELAEDVIGGAAIDKHGVPLADQTLQRARQADAVLLGAVGGPKWDRIERDIRPERGLLKIRSQLGLFANLRPAILYPQLAAASSLKPEVVAGLDILIVRELTGGIYFGQPREQRVLENGERQAYDTLPYSESEIRRIARVGFDMARVRNNRLCSVDKANVLASSQLWREVVEEVAKDYPDVELSHMYVDNAAMQLVRAPKQFDVMVTDNMFGDILSDEASMLTGSIGMLPSASLDANNKGMYEPCHGSAPDIAGQGIANPLATILSVSMMLRYSFSQATAADAIEQAVSKVLDQGLRTGDIWSEGCRKVGTREMGDAVVAALKNL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3118 and its homologs
|