| Identification |
| Name: |
Alpha-ketoglutarate-dependent taurine dioxygenase |
| Synonyms: |
- 2-aminoethanesulfonate dioxygenase
- Sulfate starvation-induced protein 3
- SSI3
|
| Gene Name: |
tauD |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in oxidoreductase activity |
| Specific Function: |
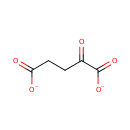
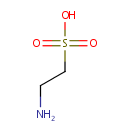
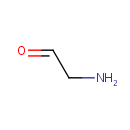
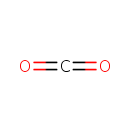
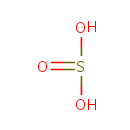
Catalyzes the conversion of taurine and alpha ketoglutarate to sulfite, aminoacetaldehyde and succinate. Required for the utilization of taurine (2-aminoethanesulfonic acid) as an alternative sulfur source. Pentane-sulfonic acid, 3- (N-morpholino)propanesulfonic acid and 1,3-dioxo-2- isoindolineethanesulfonic acid are also substrates for this enzyme |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | oxidoreductase activity | | Process |
|---|
| metabolic process | | oxidation reduction |
|
| Gene Properties |
| Locus tag: |
PA3935 |
| Strand: |
- |
| Entrez Gene ID: |
878589 |
| Accession: |
NP_252624.1 |
| GI: |
15599130 |
| Sequence start: |
4413192 |
| Sequence End: |
4414025 |
| Sequence Length: |
833 |
| Gene Sequence: |
>PA3935
ATGAGCCTGACCATCCAACCCATCAGCCCGGCCCTGGGCGCCATCGTTTCCGGCATCGACCTCGGCGCGCCGCTGGACGACACCGGCCAGCGGGCCATCGAGCAGGCCCTGCTCGAACACCAGGTGCTGTTCTTCCGCGACCAGTCGCTGGAACCCAGGAGCCAGGCGCGCTTCGCCGCCCGCTTCGGCGACCTGCACATCCATCCTATCTACCCCAGCGTGCCGGAGCAGCCGGAGGTCATCGTCCTCGACACCGCGGTCACCGACGTGCGCGACAATGCCATCTGGCACACCGACGTGACCTTCCTCGAGACCCCGGCGCTGGGAGCCGTGCTCGCCGCCAAGCAATTGCCGCCGTACGGCGGCGACACCCTCTGGGCGAGCAGCACGGCGGCCTACGAGGCGCTGTCGGCGCCGCTGCGGCGTCTGCTGGACGGCCTCACGGCGACCCACGACATCGGCAAGTCCTTCCCCCGCGAGCGCTTCGGCGTCACCGAGGCCGACCTGGCGCGCCTGGAGGAGGCCCGCCTGAAGAATCCGCCGCGCTCGCATCCGGTGGTGCGCACCCATCCGGTCACCGGGCGCAAGGGCCTGTTCGTCAGCGATGGCTTCACCACCCGCATCAACGAACTGGAGCCGGCCGAGAGCGATGCGCTGCTGAAGTTCCTCTTCGCCCATGCCACGCGACCTGAATTCACCGTGCGCTGGCGCTGGCAGGAAAACGACGTGGCGTTCTGGGACAACCGCGTGACGCAGCACTACGCGGTGGACGATTACCGGCCACAGCGGCGGGTCATGCATCGCGCCACCATCCTCGGCGACAAGCCCTTCTGA |
| Protein Properties |
| Protein Residues: |
277 |
| Protein Molecular Weight: |
31 kDa |
| Protein Theoretical pI: |
6.35 |
| Hydropathicity (GRAVY score): |
-0.287 |
| Charge at pH 7 (predicted): |
-3.59 |
| Protein Sequence: |
>PA3935
MSLTIQPISPALGAIVSGIDLGAPLDDTGQRAIEQALLEHQVLFFRDQSLEPRSQARFAARFGDLHIHPIYPSVPEQPEVIVLDTAVTDVRDNAIWHTDVTFLETPALGAVLAAKQLPPYGGDTLWASSTAAYEALSAPLRRLLDGLTATHDIGKSFPRERFGVTEADLARLEEARLKNPPRSHPVVRTHPVTGRKGLFVSDGFTTRINELEPAESDALLKFLFAHATRPEFTVRWRWQENDVAFWDNRVTQHYAVDDYRPQRRVMHRATILGDKPF |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3935 and its homologs
|