| Identification |
| Name: |
Formate dehydrogenase, nitrate-inducible, cytochrome b556(fdn) subunit |
| Synonyms: |
- Anaerobic formate dehydrogenase cytochrome b556 subunit
- Formate dehydrogenase-N subunit gamma
- FDH-N subunit gamma
|
| Gene Name: |
fdnI |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in respiratory electron transport chain |
| Specific Function: |
Formate dehydrogenase allows E.coli to use formate as major electron donor during anaerobic respiration, when nitrate is used as electron acceptor. Subunit gamma is the cytochrome b556(FDN) component of the formate dehydrogenase |
| Cellular Location: |
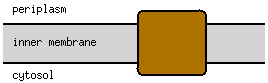
Cell inner membrane; Multi-pass membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
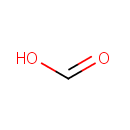
| | | formate [inner membrane] + NAD +[inner membrane] → CO 2[inner membrane] + NADH [inner membrane] + H +[inner membrane]ReactionCard | |
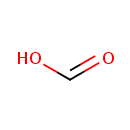 | + | ETR-Quinones | + |  | → | 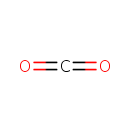 | + | ETR-Quinols |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | formate dehydrogenase complex | | integral to membrane | | intrinsic to membrane | | macromolecular complex | | membrane | | membrane part | | protein complex | | Function |
|---|
| catalytic activity | | electron carrier activity | | formate dehydrogenase activity | | oxidoreductase activity | | oxidoreductase activity, acting on the aldehyde or oxo group of donors | | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | | Process |
|---|
| cellular metabolic process | | cellular respiration | | electron transport chain | | energy derivation by oxidation of organic compounds | | generation of precursor metabolites and energy | | metabolic process | | respiratory electron transport chain |
|
| Gene Properties |
| Locus tag: |
PA4810 |
| Strand: |
- |
| Entrez Gene ID: |
880192 |
| Accession: |
NP_253498.1 |
| GI: |
15600004 |
| Sequence start: |
5396992 |
| Sequence End: |
5397618 |
| Sequence Length: |
626 |
| Gene Sequence: |
>PA4810
ATGAAAAATGAAATCCAGCGCTACAACGCCAACGAGCGCAGCAACCACTGGGCCGTGGCGATCCTGTTCTTCATGGCCGGGCTGTCCGGGCTGGCGCTGTTCCACCCGGCGCTGTTCTGGCTGACCAACCTGTTCGGCGGCGGTCCCTGGACGCGTATCCTGCACCCCTTCCTGGGGCTGGCGATGTTCGTCTTCTTCCTCGGCCTGGTGGTGCGCTTCGCCCACCACAACCGGGTGCAGAAGAGCGATATCCAGTGGCTCAAGCAGTGGCGCGACGTGGTCAGCAACCGCGAGGAGAACCTCCCGGAAGTCGGCCGCTACAACGCCGGGCAGAAGCTGCTGTTCTGGGTGCTGCTGCTGAGCATGCTGACCCTGCTGGTCACCGGCATCGTGATCTGGCGCCAGTACTTCAGTGCCTGGTTCGGCATCGAGGCGATCCGCCTGTCGGCGCTCCTGCACGCCTTCGCCGCCTTCGTCCTGATCGCCAGCATCATCGTGCACATCTACGCCGGCATCTGGGTCAAGGGCTCGATGGGCGCGATGCTCTACGGCAAGGTCAGCCGCGCCTGGGCGCGCAAGCACCATAACGGCTGGCTGAAGGAAGTCGGCAAGGGCGAGGAGCACTGA |
| Protein Properties |
| Protein Residues: |
208 |
| Protein Molecular Weight: |
23.9 kDa |
| Protein Theoretical pI: |
10.81 |
| Hydropathicity (GRAVY score): |
0.313 |
| Charge at pH 7 (predicted): |
12.39 |
| Protein Sequence: |
>PA4810
MKNEIQRYNANERSNHWAVAILFFMAGLSGLALFHPALFWLTNLFGGGPWTRILHPFLGLAMFVFFLGLVVRFAHHNRVQKSDIQWLKQWRDVVSNREENLPEVGRYNAGQKLLFWVLLLSMLTLLVTGIVIWRQYFSAWFGIEAIRLSALLHAFAAFVLIASIIVHIYAGIWVKGSMGAMLYGKVSRAWARKHHNGWLKEVGKGEEH |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4810 and its homologs
|