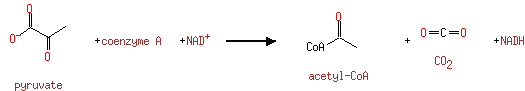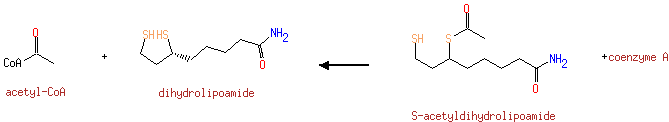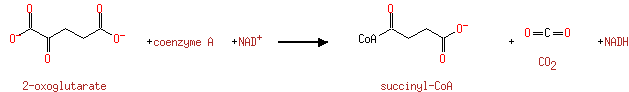| Identification |
| Name: |
2-oxoglutarate dehydrogenase E1 component |
| Synonyms: |
- Alpha-ketoglutarate dehydrogenase
|
| Gene Name: |
sucA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in oxoglutarate dehydrogenase (succinyl-transferring) activity |
| Specific Function: |
The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components:2- oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3) |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
| | |
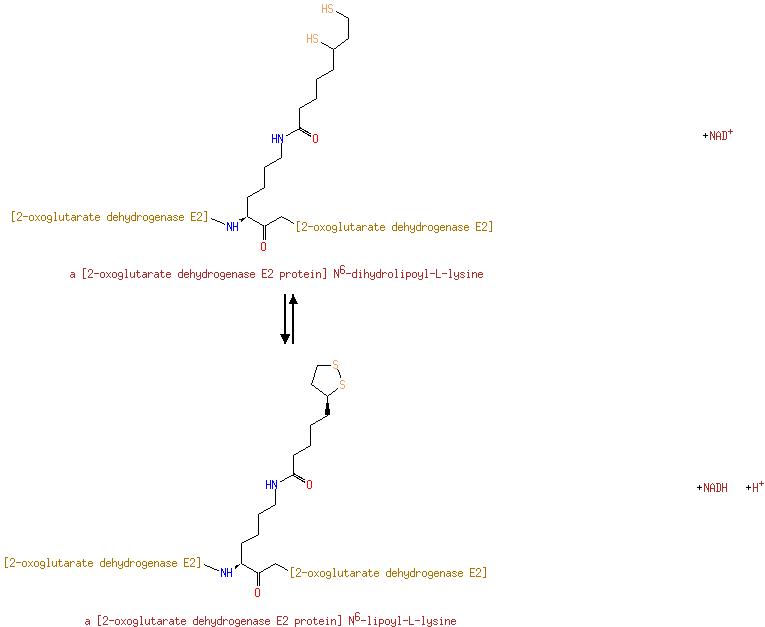
 | + | Enzyme N6-(lipoyl)lysine | ↔ | [Dihydrolipoyllysine-residue succinyltransferase] S-glutaryldihydrolipoyllysine | + |  |
| | |
 | ↔ | [Dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine | + |  |
| |
|
| SMPDB Reactions: |
| | |
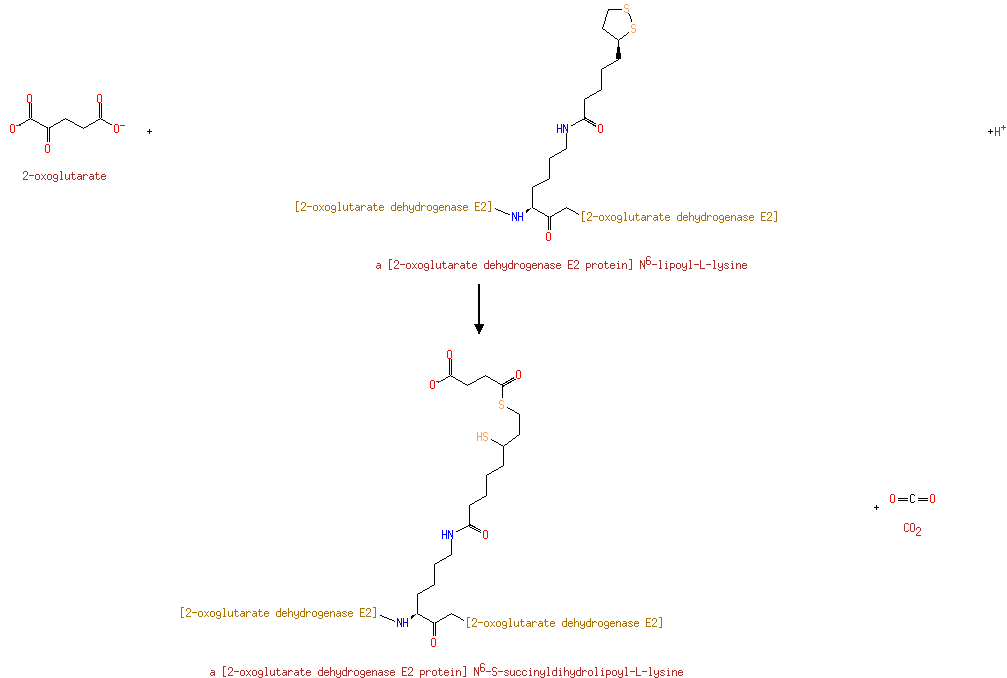
a [2-oxoglutarate dehydrogenase E2 protein] N6-lipoyl-L-lysine | + | 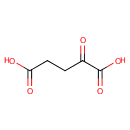 | + |  | → | a [2-oxoglutarate dehydrogenase E2 protein] N6-S-succinyldihydrolipoyl-L-lysine | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
| | |
 | + | Enzyme N6-(lipoyl)lysine | ↔ | [Dihydrolipoyllysine-residue succinyltransferase] S-glutaryldihydrolipoyllysine | + |  |
| | |
 | + | [dihydrolipoyllysine-residue succinyltransferase] lipoyllysine | → | [dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine | + |  |
| | |
 | ↔ | [Dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine | + |  |
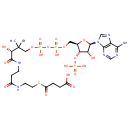
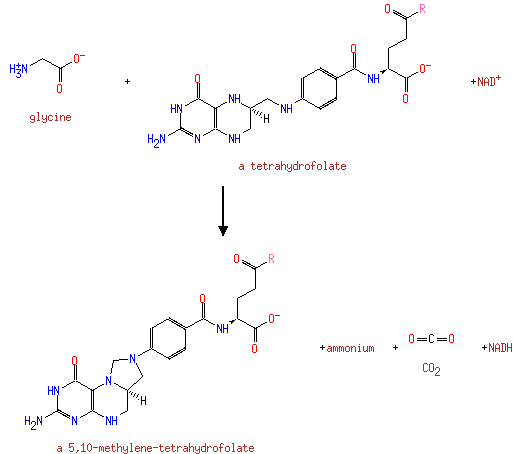
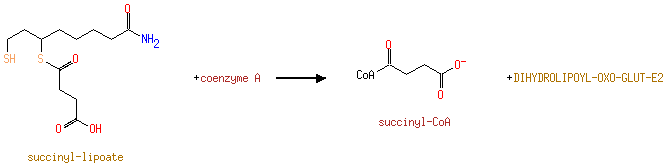
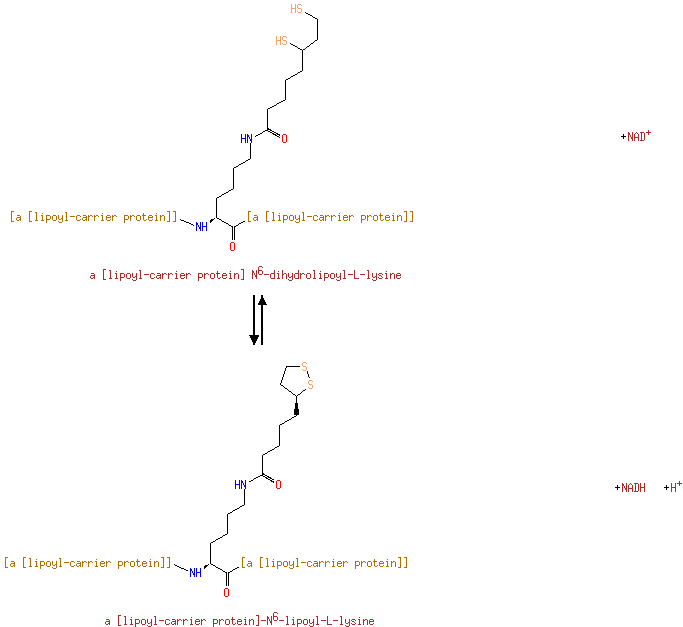
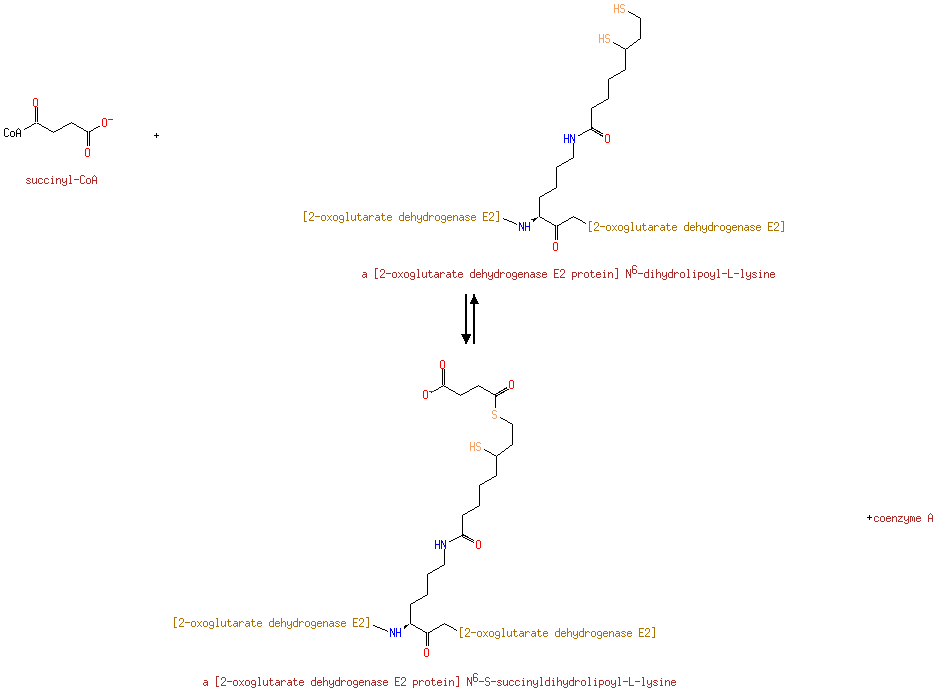
| | | | | glycine + a tetrahydrofolate + NAD + → a 5,10-methylene-tetrahydrofolate + ammonium + CO 2 + NADH ReactionCard | | pyruvate + coenzyme A + NAD + → acetyl-CoA + CO 2 + NADH ReactionCard | | pyruvate + lipoamide → S-acetyldihydrolipoamide + CO 2ReactionCard | | acetyl-CoA + dihydrolipoamide ← S-acetyldihydrolipoamide + coenzyme A ReactionCard | | 2-oxoglutarate + coenzyme A + NAD + → succinyl-CoA + CO 2 + NADH ReactionCard | | a [glycine-cleavage complex H protein] N6-aminomethyldihydrolipoyl-L-lysine + a tetrahydrofolate ↔ a [glycine-cleavage complex H protein] N6-dihydrolipoyl-L-lysine + a 5,10-methylene-tetrahydrofolate + ammonium ReactionCard | | glycine + a [glycine-cleavage complex H protein] N6-lipoyl-L-lysine + H + ↔ a [glycine-cleavage complex H protein] N6-aminomethyldihydrolipoyl-L-lysine + CO 2ReactionCard | | succinyl-lipoate + coenzyme A → succinyl-CoA + DIHYDROLIPOYL-OXO-GLUT-E2 ReactionCard | | a [lipoyl-carrier protein] N6-dihydrolipoyl-L-lysine + NAD + = a [lipoyl-carrier protein]- N6-lipoyl-L-lysine + NADH + H +ReactionCard | | 2-oxoglutarate + a [2-oxoglutarate dehydrogenase E2 protein] N6-lipoyl-L-lysine + H + → a [2-oxoglutarate dehydrogenase E2 protein] N6- S-succinyldihydrolipoyl-L-lysine + CO 2ReactionCard | | succinyl-CoA + a [2-oxoglutarate dehydrogenase E2 protein] N6-dihydrolipoyl-L-lysine = a [2-oxoglutarate dehydrogenase E2 protein] N6- S-succinyldihydrolipoyl-L-lysine + coenzyme A ReactionCard | | a [2-oxoglutarate dehydrogenase E2 protein] N6-dihydrolipoyl-L-lysine + NAD + = a [2-oxoglutarate dehydrogenase E2 protein] N6-lipoyl-L-lysine + NADH + H +ReactionCard |
|
| Complex Reactions: |
| | |
 | + | [dihydrolipoyllysine-residue succinyltransferase] lipoyllysine | → | [dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine | + |  |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | oxidoreductase activity | | oxidoreductase activity, acting on the aldehyde or oxo group of donors | | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor | | oxoglutarate dehydrogenase (succinyl-transferring) activity | | thiamin pyrophosphate binding | | vitamin binding | | Process |
|---|
| alcohol metabolic process | | glucose catabolic process | | glucose metabolic process | | glycolysis | | hexose metabolic process | | metabolic process | | monosaccharide metabolic process | | small molecule metabolic process |
|
| Gene Properties |
| Locus tag: |
PA1585 |
| Strand: |
+ |
| Entrez Gene ID: |
881879 |
| Accession: |
NP_250276.1 |
| GI: |
15596782 |
| Sequence start: |
1724244 |
| Sequence End: |
1727075 |
| Sequence Length: |
2831 |
| Gene Sequence: |
>PA1585
ATGCACGAAAGCGTAATGCAGCGGATGTGGAACAGTGCCCATCTATCCGGTGGTAACGCTGCCTACGTCGAAGAGCTCTACGAGCTCTACCTGCACGACCCTAACGCTGTGCCAGAAGAGTGGCGCACGTATTTCCAGAAGCTTCCCGCCGATGGCAATCCCGCCCCCGACGTCTCGCACTCCACAGTGCGCGATCATTTCGTCCTGCTGGCCAAGAACCAGCGCCGTGCCCAGCCGGTTTCCGCCGGTAGCGTGAGCAGCGAGCACGAGAAGAAGCAGGTCGAAGTCCTTCGTCTCATTCACGCTTACCGTCTCCGCGGCCACCAGGCTTCCACCCTCGACCCGCTCGGCCTGTGGCAGCGTCCCGCGCCTGCCGACCTGTCGATCGACCACTACGGTCTGACGGGTGCCGACCTGGACACCACTTTCCGCACCGGTGAGCTTTACATCGGCAAGGAAGAGGCGACCCTGCGTGAAATCGTCGACAGCCTGAAGAGCACCTACTGCGGTACCTTCGGCGCCGAGTTCATGCACATCGTCGATTCCGAGCAGCGCAAGTGGTTCCTCCAGCGTCTGGAAAGCGTGCGTGGTCGTCCCGGCTTCTCCGCCGAGGCGCGTGCGCACCTGCTCGAGCGCCTGACCGCGGCCGAAGGCCTGGAGAAGTACCTGGGCACCAAGTACCCGGGCACCAAGCGTTTCGGCCTGGAAGGCGGCGAAAGCCTGATCCCGATGGTCGACGAGATCATCCAGCGCTGCGGTTCCTACGGCGCCAAGGAAATCGTCATCGGCATGGCCCACCGCGGTCGCCTGAACGTCCTGGTCAACACCCTGGGCAAGAACCCGCGCGACCTGTTCGACGAGTTCGAAGGCAAGAAGATCGTCGAGCTGGGCTCCGGTGACGTGAAGTACCACCAGGGCTTCTCCTCGAACGTGATGACCAGCGGTGGCGAAGTCCACCTGGCGCTGGCGTTCAACCCGTCCCACCTGGAAATCGTTTCCCCGGTGGTCGAGGGTTCCGTTCGTGCTCGCCAGGACCGCCGCAAGGACAGCTCCGGCGACAAGGTGGTACCGATTTCCATCCACGGCGATGCCGCCTTCGCGGGGCAGGGCGTGGTCATGGAAACCTTCCAGATGTCGCAGACCCGCGCCTACAAGACCGGTGGCACCATCCACCTGGTGATCAACAACCAGGTCGGTTTCACCACCAGCCGCCAGGACGATGCGCGTTCCACCGAGTACGCCACCGACGTGGCGAAGATGATTCAGGCGCCGATCTTCCACGTCAACGGCGACGATCCCGAAGCCGTGCTGTTCGTCACCCAACTGGCCGTCGACTACCGCATGCAGTTCAAGCGTGACGTGGTCATCGACCTGGTCTGCTACCGTCGCCGCGGCCACAACGAGGCGGACGAGCCGAGCGGCACCCAGCCGCTGATGTACCAGCAGATCGCCAAGCAGCGTACTACCCGCGAGCTGTACGCCGATGCCCTGGTGAATGCCGGCGTGCTCAGCGCCGAGCAGGTTCAGTCGAAGATCGACGACTACCGCGACGCCCTTGACAACGGCCTGCACGTGGTGAAGAGCCTGGTCAAGGAACCCAACAAGGAACTGTTCGTCGACTGGCGTCCGTACCTGGGTCATGCCTGGACCGCGCGTCACGACACCCGCTTCGACCTGAAGACCCTGCAGGAACTGTCCAGCAAGATGCTGGAAGTGCCGGAAGGCTTCGTGGTTCAGCGCCAGGTTTCGAAGATCTACGAAGATCGCCAGAAGATGGCGGCCGGTGGCCTGCCGATCAACTGGGGCTTCGCCGAGACCCTGGCCTACGCGACCCTGCTGTTCGAAGGCCATCCGGTACGGATGACCGGCCAGGACGTCGGCCGTGGCACCTTCTCGCACCGTCACGCGGTGCTGCACAACCAGAAGGATGACTCGGTCTACGTACCGCTGGCCAATCTGTTCGACGGCCAGCCGCGCCTGGATATCTACGATTCCTTCCTTTCGGAAGAGGCCGTGCTGGCGTTCGAATACGGTTTCGCCACCACCACGCCGAACTCGCTGGTGATCTGGGAAGCCCAGTTCGGCGACTTCGCCAACGGCGCCCAGGTGGTCATCGACCAGTTCATCACCAGTGGCGAAAGCAAATGGGGCCGACTCTGCGGCCTGACCATGCTGCTGCCGCACGGCTATGAGGGGCAGGGCCCGGAGCACTCCTCCGCGCGCCTGGAGCGCTACCTGCAGCTCTGTGCCGAGCAGAACATCCAGGTCTGCGTGCCGACCACCCCGGCTCAGGTCTACCACATGCTCCGCCGTCAGGTGATCCGCCCGCTGCGCAAGCCGCTGGTGGTGATGACGCCGAAGTCCCTGCTGCGTCACAAGCTCGCCATCTCGACCCTGGAAGATCTGGCCAACGGTTCCTTCCAGACTGTGATCCCGGAAATCGACAGCCTGGATCCGAAGAAGGTCGACCGCGTCGTGCTGTGCAGCGGCAAGGTGTACTACGACCTGCTGGAGAAGCGTCGCGCGGAAGGCCGCGAAGATACCGCGATCGTCCGTATCGAGCAGCTGTATCCGTTCCCGGAAGACGACCTGGCGGAAGTTCTGGCTCCGTACAAGAACCTCAAGCACATCGTCTGGTGCCAGGAAGAGCCGATGAACCAGGGCGCCTGGTTCTGCAGCCAGCATCACATGCGTCGTGTCATCGCCGCGCACAAGAAGGGCCTCAATCTCGAGTACGCCGGCCGCGAAGGCTCCGCTGCTCCGGCTTGCGGCTACGCCTCGATGCACGCCGAGCAGCAGGAAAAACTGCTGCAAGACGCCTTTACTGTTTAA |
| Protein Properties |
| Protein Residues: |
943 |
| Protein Molecular Weight: |
105.9 kDa |
| Protein Theoretical pI: |
6.54 |
| Hydropathicity (GRAVY score): |
-0.378 |
| Charge at pH 7 (predicted): |
-8.83 |
| Protein Sequence: |
>PA1585
MHESVMQRMWNSAHLSGGNAAYVEELYELYLHDPNAVPEEWRTYFQKLPADGNPAPDVSHSTVRDHFVLLAKNQRRAQPVSAGSVSSEHEKKQVEVLRLIHAYRLRGHQASTLDPLGLWQRPAPADLSIDHYGLTGADLDTTFRTGELYIGKEEATLREIVDSLKSTYCGTFGAEFMHIVDSEQRKWFLQRLESVRGRPGFSAEARAHLLERLTAAEGLEKYLGTKYPGTKRFGLEGGESLIPMVDEIIQRCGSYGAKEIVIGMAHRGRLNVLVNTLGKNPRDLFDEFEGKKIVELGSGDVKYHQGFSSNVMTSGGEVHLALAFNPSHLEIVSPVVEGSVRARQDRRKDSSGDKVVPISIHGDAAFAGQGVVMETFQMSQTRAYKTGGTIHLVINNQVGFTTSRQDDARSTEYATDVAKMIQAPIFHVNGDDPEAVLFVTQLAVDYRMQFKRDVVIDLVCYRRRGHNEADEPSGTQPLMYQQIAKQRTTRELYADALVNAGVLSAEQVQSKIDDYRDALDNGLHVVKSLVKEPNKELFVDWRPYLGHAWTARHDTRFDLKTLQELSSKMLEVPEGFVVQRQVSKIYEDRQKMAAGGLPINWGFAETLAYATLLFEGHPVRMTGQDVGRGTFSHRHAVLHNQKDDSVYVPLANLFDGQPRLDIYDSFLSEEAVLAFEYGFATTTPNSLVIWEAQFGDFANGAQVVIDQFITSGESKWGRLCGLTMLLPHGYEGQGPEHSSARLERYLQLCAEQNIQVCVPTTPAQVYHMLRRQVIRPLRKPLVVMTPKSLLRHKLAISTLEDLANGSFQTVIPEIDSLDPKKVDRVVLCSGKVYYDLLEKRRAEGREDTAIVRIEQLYPFPEDDLAEVLAPYKNLKHIVWCQEEPMNQGAWFCSQHHMRRVIAAHKKGLNLEYAGREGSAAPACGYASMHAEQQEKLLQDAFTV |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA1585 and its homologs
|