| Identification |
| Name: |
Uroporphyrinogen decarboxylase |
| Synonyms: |
|
| Gene Name: |
hemE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in uroporphyrinogen decarboxylase activity |
| Specific Function: |
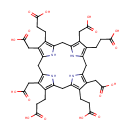
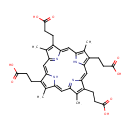
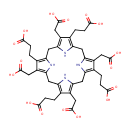
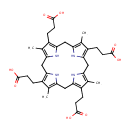
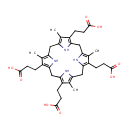
Catalyzes the decarboxylation of four acetate groups of uroporphyrinogen-III to yield coproporphyrinogen-III |
| Cellular Location: |
Cytoplasm (Probable) |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| carbon-carbon lyase activity | | carboxy-lyase activity | | catalytic activity | | lyase activity | | uroporphyrinogen decarboxylase activity | | Process |
|---|
| metabolic process | | nitrogen compound metabolic process | | porphyrin biosynthetic process | | porphyrin metabolic process | | tetrapyrrole metabolic process |
|
| Gene Properties |
| Locus tag: |
PA5034 |
| Strand: |
- |
| Entrez Gene ID: |
881202 |
| Accession: |
NP_253721.1 |
| GI: |
15600227 |
| Sequence start: |
5664990 |
| Sequence End: |
5666057 |
| Sequence Length: |
1067 |
| Gene Sequence: |
>PA5034
ATGACTGCCCTGAAGAACGACCGCTTCCTCCGCGCCCTGCTCAAGCAGCCCGTCGATGTCACCCCCGTCTGGATGATGCGCCAGGCCGGTCGTTACCTGCCGGAATACCGTGCCACCCGTGCCAAGGCCGGCGATTTCATGAGCCTGTGCATGAACCCGGAGCTGGCCTGCGAAGTGACCCTGCAGCCGCTGGACCGTTACCCGCAGCTGGACGCCGCGATCCTCTTCTCCGATATCCTGACCATTCCCGATGCCATGGGCCAGGGCCTGTACTTCGAGACCGGCGAAGGCCCGCGCTTCCGCAAGGTGGTCTCGTCGCTGGCCGACATCGAGGCGCTGCCGGTTCCCGATCCGGAACAGGACCTGGGCTATGTGATGGACGCCGTGCGCACCATCCGCCGCGAGCTGAACGGCCGCGTACCGCTGATCGGCTTCTCCGGCAGCCCCTGGACCCTCGCCACCTACATGGTCGAGGGCGGCTCGAGCAAGGACTTCCGCAAGTCCAAGGCGATGCTCTACGACAACCCCAAGGCCATGCACGCTCTGCTGGACAAGCTGGCGCAATCGGTGACCAGCTACCTCAACGGGCAGATCCACGCCGGCGCCCAGGCGGTGCAGATCTTCGACTCATGGGGCGGCAGCCTGTCGGCGGCGGCCTACCAGGAGTTCTCCCTGGCCTACATGCGCAAGATCGTCGATGGATTGATCCGCGAGCACGACGGACGCCGGGTGCCGGTGATCCTCTTCACCAAGGGAGGCGGCCTGTGGCTGGAGTCGATGGCCGAGGTCGGCGCCGAGGCGCTGGGCCTGGACTGGACCTGCGACATCGGCAGCGCCCGGGCCCGTGTCGGTGAGCGCGTAGCCCTGCAAGGCAACATGGATCCTTCGGTGCTCTACGCCAATCCGGCGGCGATCCGCGCCGAAGTCGCGCGGATCCTGGCGGCCTACGGCAAGGGTACGGGGCATGTGTTCAATCTCGGCCACGGCATCACCCCGGAAGTCGACCCGGCCCATGCCGGCGCCTTCTTCGAGGCGGTGCACGAGCTGTCGGCGCAGTACCACGGCTGA |
| Protein Properties |
| Protein Residues: |
355 |
| Protein Molecular Weight: |
38.8 kDa |
| Protein Theoretical pI: |
6.32 |
| Hydropathicity (GRAVY score): |
-0.069 |
| Charge at pH 7 (predicted): |
-3.17 |
| Protein Sequence: |
>PA5034
MTALKNDRFLRALLKQPVDVTPVWMMRQAGRYLPEYRATRAKAGDFMSLCMNPELACEVTLQPLDRYPQLDAAILFSDILTIPDAMGQGLYFETGEGPRFRKVVSSLADIEALPVPDPEQDLGYVMDAVRTIRRELNGRVPLIGFSGSPWTLATYMVEGGSSKDFRKSKAMLYDNPKAMHALLDKLAQSVTSYLNGQIHAGAQAVQIFDSWGGSLSAAAYQEFSLAYMRKIVDGLIREHDGRRVPVILFTKGGGLWLESMAEVGAEALGLDWTCDIGSARARVGERVALQGNMDPSVLYANPAAIRAEVARILAAYGKGTGHVFNLGHGITPEVDPAHAGAFFEAVHELSAQYHG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5034 and its homologs
|