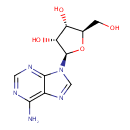
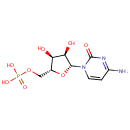
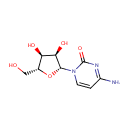
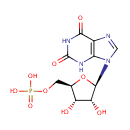
| Identification |
| Name: |
acetylpolyamine aminohydrolase |
| Synonyms: |
Not Available |
| Gene Name: |
aphA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
putrescine biosynthetic process, polyamine metabolic process, hydrolase activity, amidase activity |
| Specific Function: |
acetylpolyamine amidohydrolase, involved in the conversion of agmatine into putrescine. |
| Cellular Location: |
Periplasm (Potential), Cytoplasmic |
| KEGG Pathways: |
- 1,4-Dichlorobenzene degradation pae00627
- Microbial metabolism in diverse environments pae01120
- Riboflavin metabolism pae00740
- gamma-Hexachlorocyclohexane degradation pae00361
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |
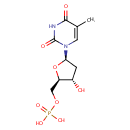
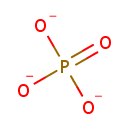
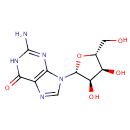
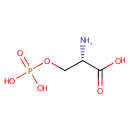
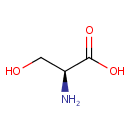
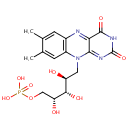
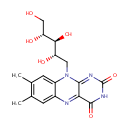
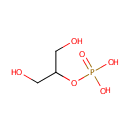
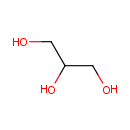
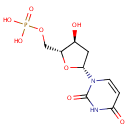
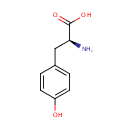
A phosphate monoester | + |  | → | an alcohol | + | 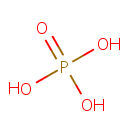 |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | outer membrane-bounded periplasmic space | | periplasmic space | | Function |
|---|
| acid phosphatase activity | | catalytic activity | | hydrolase activity | | hydrolase activity, acting on ester bonds | | phosphatase activity | | phosphoric ester hydrolase activity |
|
| Gene Properties |
| Locus tag: |
PA1409 |
| Strand: |
+ |
| Entrez Gene ID: |
879295 |
| Accession: |
NP_250100.1 |
| GI: |
15596606 |
| Sequence start: |
1533238 |
| Sequence End: |
1534278 |
| Sequence Length: |
1040 |
| Gene Sequence: |
>PA1409
ATGCTGAGCGTCTACAGCGACGATCATCGTCTGCACTTCGGCCAGTCGGAACTGGTCGACGGCAAGCTGCAACCCTGTTTCGAAATGCCCAGCCGGGCCGACACCGTGCTGGCCCGGGTGAAGTCGCAGAACCTTGGCGAGGTGATCGCGCCGAAGGATTTCGGCCGCGAGCCGCTGCTGCGCCTCCACGACGCCGCCTACCTGGACTTCCTCCAGGGCGCCTGGGCCCGCTGGACCGCCGAAGGCCACAGCGGCGACCTGGTGTCCACCACCTTCCCCGGCCGCCGCCTGCGCCGCGACGGGCCGATCCCCACGGCGCTGATGGGCGAGCTGGGCTACTACAGCTTCGACACCGAGGCGCCGATCACCGCCGGCACCTGGCAGGCCATCTACAGCTCCGCCCAGGTCGCGCTGACCGCCCAGGAGCATATGCGCCAGGGCGCCCGCAGCGCCTTCGCCCTCTGCCGTCCGCCGGGCCATCACGCCGGTGGCGACTTCATGGGCGGCTACTGCTTCCTCAACAATGCCGCCATCGCCACCCAGGCGTTCCTCGACCAGGGTGCCCGGCGCGTGGCGATCCTCGATGTCGACTATCACCATGGCAACGGCACCCAGGACATTTTCTACCGCCGCGACGATGTGCTGTTCGCCTCGATCCATGGCGATCCGCGGGTCGAATACCCGTACTTCCTCGGCTACGCAGACGAGCGCGGCGAAGGCGCCGGCGAGGGCTGCAACCACAACTATCCGCTGGCCCATGGCAGCGGCTGGGACCTCTGGTCGGCGGCGCTCGACGACGCCTGCGTGCGGATCGCCGGCTACGCCCCGGATGCCCTGGTGATTTCCCTTGGCGTGGACACCTACAAGGAAGACCCGATCTCCCAGTTCAGGCTGGATTCGCCGGACTACCTGCGGATGGGCGAGCGCATCGCCCGGCTGGGCCTGCCGACCCTGTTCATCATGGAAGGCGGCTATGCGGTGGAAGCCATCGGCATCAACGCGGTCAACGTGCTGCAGGGCTACGAAGGCGCGGCCCGCTGA |
| Protein Properties |
| Protein Residues: |
346 |
| Protein Molecular Weight: |
38 kDa |
| Protein Theoretical pI: |
5.07 |
| Hydropathicity (GRAVY score): |
-0.231 |
| Charge at pH 7 (predicted): |
-12.27 |
| Protein Sequence: |
>PA1409
MLSVYSDDHRLHFGQSELVDGKLQPCFEMPSRADTVLARVKSQNLGEVIAPKDFGREPLLRLHDAAYLDFLQGAWARWTAEGHSGDLVSTTFPGRRLRRDGPIPTALMGELGYYSFDTEAPITAGTWQAIYSSAQVALTAQEHMRQGARSAFALCRPPGHHAGGDFMGGYCFLNNAAIATQAFLDQGARRVAILDVDYHHGNGTQDIFYRRDDVLFASIHGDPRVEYPYFLGYADERGEGAGEGCNHNYPLAHGSGWDLWSAALDDACVRIAGYAPDALVISLGVDTYKEDPISQFRLDSPDYLRMGERIARLGLPTLFIMEGGYAVEAIGINAVNVLQGYEGAAR |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA1409 and its homologs
|