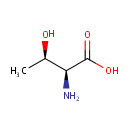
L-Threonine (PAMDB000068)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000068 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Threonine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Threonine is an amino acid. It is converted to pyruvate via threonine dehydrogenase. An intermediate in this pathway can undergo thiolysis with CoA to produce acetyl-CoA and glycine. (Wikipedia) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C4H9NO3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 119.1192 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 119.058243159 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | AYFVYJQAPQTCCC-GBXIJSLDSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C4H9NO3/c1-2(6)3(5)4(7)8/h2-3,6H,5H2,1H3,(H,7,8)/t2-,3+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 72-19-5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S,3R)-2-amino-3-hydroxybutanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-threonine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[C@@H](O)[C@H](N)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acids and conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hydroxy fatty acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 256 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Threonine <> Acetaldehyde + Glycine L-Threonine > 2-Ketobutyric acid + Ammonium Adenosine triphosphate + Water + L-Threonine > ADP + Hydrogen ion + Phosphate + L-Threonine Adenosine triphosphate + Water + L-Threonine > ADP + Hydrogen ion + Phosphate + L-Threonine Water + O-Phosphohomoserine <> Phosphate + L-Threonine Adenosine triphosphate + L-Threonine + tRNA(Thr) + tRNA(Thr) <> Adenosine monophosphate + Pyrophosphate + L-Threonyl-tRNA(Thr) + L-Threonyl-tRNA(Thr) NAD + L-Threonine <> L-2-Amino-3-oxobutanoic acid + Hydrogen ion + NADH Water + L-Threonine O-3-phosphate > Phosphate + L-Threonine L-Threonine <> 2-Ketobutyric acid + Ammonia Adenosine triphosphate + L-Threonine + tRNA(Thr) <> Adenosine monophosphate + Pyrophosphate + L-Threonyl-tRNA(Thr) ala-thr + Water > L-Alanine + L-Threonine L-Threonine > Hydrogen ion + 2-Ketobutyric acid + Ammonia L-Threonine + NAD > L-2-Amino-3-oxobutanoic acid + NADH O-Phosphohomoserine + Water > L-Threonine + Inorganic phosphate L-Threonine + 2-Aminobut-2-enoate + 2-Iminobutanoate + Water <> 2-Ketobutyric acid + Ammonia L-Threonine + Adenosine triphosphate + Hydrogen carbonate <> L-Threonylcarbamoyladenylate + Pyrophosphate + Water More...L-Threonine + Adenosine triphosphate + Hydrogen ion + tRNA(Thr) + L-Threonine > Pyrophosphate + Adenosine monophosphate + L-Threonyl-tRNA(Thr) O-Phosphohomoserine + Water > Phosphate + L-Threonine + L-Threonine L-Threonine + L-Threonine > Hydrogen ion + Water + (2Z)-2-aminobut-2-enoate L-Threonine + NAD + L-Threonine > Hydrogen ion + NADH + L-2-Amino-3-oxobutanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Fujita, Chuzo; Nara, Takashi; Samejima, Hirotoshi; Kinoshita, Shukuo. L-Threonine fermentation. I. Microbial conversion of L-homoserine to L-threonine. Nippon Nogei Kagaku Kaishi (1965), 39(6), 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the gamma-elimination of phosphate from L- phosphohomoserine and the beta-addition of water to produce L- threonine. To a lesser extent, is able to slowly catalyze the deamination of L-threonine into alpha-ketobutyrate and that of L- serine and 3-chloroalanine into pyruvate. Is also able to rapidly convert vinylglycine to threonine, which proves that the pyridoxal p-quinonoid of vinylglycine is an intermediate in the TS reaction
- Gene Name:
- thrC
- Locus Tag:
- PA3735
- Molecular weight:
- 51.8 kDa
Reactions
| O-phospho-L-homoserine + H(2)O = L-threonine + phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the formation of alpha-ketobutyrate from threonine in a two-step reaction. The first step is a dehydration of threonine, followed by rehydration and liberation of ammonia. Deaminates L-threonine, but also L-serine to a lesser extent
- Gene Name:
- ilvA
- Locus Tag:
- PA1326
- Molecular weight:
- 55.9 kDa
Reactions
| L-threonine = 2-oxobutanoate + NH(3). |
- General function:
- Involved in catalytic activity
- Specific function:
- Interconversion of serine and glycine
- Gene Name:
- glyA
- Locus Tag:
- PA4602
- Molecular weight:
- 45.2 kDa
Reactions
| 5,10-methylenetetrahydrofolate + glycine + H(2)O = tetrahydrofolate + L-serine. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ThrS is also a translational repressor protein, it controls the translation of its own gene by binding to its mRNA
- Gene Name:
- thrS
- Locus Tag:
- PA2744
- Molecular weight:
- 73.1 kDa
Reactions
| ATP + L-threonine + tRNA(Thr) = AMP + diphosphate + L-threonyl-tRNA(Thr). |
- General function:
- Involved in acid phosphatase activity
- Specific function:
- Dephosphorylates several organic phosphomonoesters and catalyzes the transfer of low-energy phosphate groups from phosphomonoesters to hydroxyl groups of various organic compounds. Preferentially acts on aryl phosphoesters. Might function as a broad-spectrum dephosphorylating enzyme able to scavenge both 3'- and 5'-nucleotides and also additional organic phosphomonoesters
- Gene Name:
- aphA
- Locus Tag:
- PA1409
- Molecular weight:
- 38 kDa
Reactions
| A phosphate monoester + H(2)O = an alcohol + phosphate. |
- General function:
- Involved in lyase activity
- Specific function:
- Catalyzes the cleavage of L-allo-threonine and L- threonine to glycine and acetaldehyde. L-threo-phenylserine and L- erythro-phenylserine are also good substrates
- Gene Name:
- ltaE
- Locus Tag:
- PA0902
- Molecular weight:
- 35.4 kDa
Reactions
| L-threonine = glycine + acetaldehyde. |
| L-allo-threonine = glycine + acetaldehyde. |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livH
- Locus Tag:
- PA1073
- Molecular weight:
- 32.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livM
- Locus Tag:
- PA1072
- Molecular weight:
- 45.6 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livG
- Locus Tag:
- PA1071
- Molecular weight:
- 28.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livF
- Locus Tag:
- PA1070
- Molecular weight:
- 25.6 kDa
- General function:
- threonylcarbamoyladenosine biosynthetic process
- Specific function:
- Required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. Catalyzes the conversion of L-threonine, bicarbonate/CO(2) and ATP to give threonylcarbamoyl-AMP (TC-AMP) as the acyladenylate intermediate, with the release of pyrophosphate. Is also able to catalyze the reverse reaction in vitro, i.e. the formation of ATP from TC-AMP and PPi. Shows higher affinity for the full-length tRNA(Thr) lacking only the t(6)A37 modification than for its fully modified counterpart. Could also be required for the maturation of 16S rRNA. Binds to double-stranded RNA but does not interact tightly with either of the ribosomal subunits, or the 70S particles.
- Gene Name:
- tsaC
- Locus Tag:
- PA0022
- Molecular weight:
- 20.4 kDa
Reactions
| L-threonine + ATP + bicarbonate = L-threonylcarbamoyladenylate + diphosphate + H(2)O |
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Probably part of a binding-protein-dependent transport system yecCS for an amino acid. Probably responsible for energy coupling to the transport system
- Gene Name:
- yecC
- Locus Tag:
- PA5152
- Molecular weight:
- 28.4 kDa
- General function:
- Involved in transport
- Specific function:
- Specific function unknown
- Gene Name:
- rhtA
- Locus Tag:
- PA1360
- Molecular weight:
- 30.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livH
- Locus Tag:
- PA1073
- Molecular weight:
- 32.5 kDa
- General function:
- Involved in sodium:dicarboxylate symporter activity
- Specific function:
- Involved in the import of serine and threonine into the cell, with the concomitant import of sodium (symport system)
- Gene Name:
- sstT
- Locus Tag:
- PA2042
- Molecular weight:
- 42.4 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livM
- Locus Tag:
- PA1072
- Molecular weight:
- 45.6 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livG
- Locus Tag:
- PA1071
- Molecular weight:
- 28.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livF
- Locus Tag:
- PA1070
- Molecular weight:
- 25.6 kDa