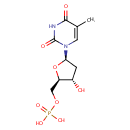
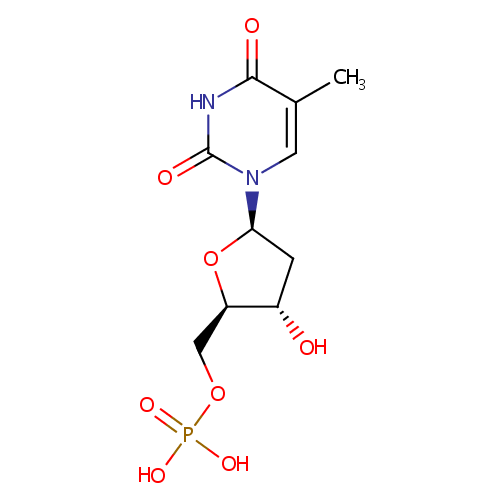
5-Thymidylic acid (PAMDB000303)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000303 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 5-Thymidylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 5-Thymidylic acid is a thymine nucleotide. Also known as thymidine monophosphate, and abbreviated TMP, is a nucleotide that is found in DNA. It is an ester of phosphoric acid with the nucleoside thymidine. TMP consists of a phosphate group, esterified to the the pentose sugar deoxyribose moiety, and the nucleobase thymine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H15N2O8P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 322.2085 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 322.056601978 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | GYOZYWVXFNDGLU-XLPZGREQSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H15N2O8P/c1-5-3-12(10(15)11-9(5)14)8-2-6(13)7(20-8)4-19-21(16,17)18/h3,6-8,13H,2,4H2,1H3,(H,11,14,15)(H2,16,17,18)/t6-,7+,8+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 365-07-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)oxolan-2-yl]methoxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | thymidylate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC1=CN([C@H]2C[C@H](O)[C@@H](COP(O)(O)=O)O2)C(=O)NC1=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as pyrimidine deoxyribonucleotides. These are pyrimidine nucleotides where the purine moiety is linked to a ribose lacking a hydroxyl group at one or more positions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrimidine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrimidine deoxyribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrimidine deoxyribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 5-Thymidylic acid + Water > Phosphate + Thymidine Thymidine 5'-triphosphate + Water > 5-Thymidylic acid + Hydrogen ion + Pyrophosphate Adenosine triphosphate + 5-Thymidylic acid <> ADP + dTDP Adenosine triphosphate + Thymidine <> ADP + 5-Thymidylic acid + Hydrogen ion dUMP + 5,10-Methylene-THF <> Dihydrofolic acid + 5-Thymidylic acid Adenosine triphosphate + Thymidine <> ADP + 5-Thymidylic acid Dihydrofolic acid + 5-Thymidylic acid + Dihydrofolic acid > 5,10-Methylene-THF + dUMP + 5,10-Methylene-THF dUMP + 5,10-methenyltetrahydrofolate mono-L-glutamate + 5,10-methenyltetrahydrofolate mono-L-glutamate > Dihydrofolic acid + 5-Thymidylic acid + Dihydrofolic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Scarano, E. Enzymic formation of thymidylic acid from 5-methyl-5'-deoxycytidylic acid. Bollettino - Societa Italiana di Biologia Sperimentale (1958), 34 945. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in thymidylate kinase activity
- Specific function:
- Catalyzes the reversible phosphorylation of deoxythymidine monophosphate (dTMP) to deoxythymidine diphosphate (dTDP), using ATP as its preferred phosphoryl donor. Situated at the junction of both de novo and salvage pathways of deoxythymidine triphosphate (dTTP) synthesis, is essential for DNA synthesis and cellular growth
- Gene Name:
- tmk
- Locus Tag:
- PA2962
- Molecular weight:
- 23.1 kDa
Reactions
| ATP + dTMP = ADP + dTDP. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Nucleotidase with a broad substrate specificity as it can dephosphorylate various ribo- and deoxyribonucleoside 5'- monophosphates and ribonucleoside 3'-monophosphates with highest affinity to 3'-AMP. Also hydrolyzes polyphosphate (exopolyphosphatase activity) with the preference for short-chain- length substrates (P20-25). Might be involved in the regulation of dNTP and NTP pools, and in the turnover of 3'-mononucleotides produced by numerous intracellular RNases (T1, T2, and F) during the degradation of various RNAs. Also plays a significant physiological role in stress-response and is required for the survival of Pseudomonas aeruginosa in stationary growth phase
- Gene Name:
- surE
- Locus Tag:
- PA3625
- Molecular weight:
- 26.4 kDa
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| A 3'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| (Polyphosphate)(n) + H(2)O = (polyphosphate)(n-1) + phosphate. |
- General function:
- Involved in thymidylate synthase activity
- Specific function:
- Provides the sole de novo source of dTMP for DNA biosynthesis. This protein also binds to its mRNA thus repressing its own translation
- Gene Name:
- thyA
- Locus Tag:
- PA0342
- Molecular weight:
- 30 kDa
Reactions
| 5,10-methylenetetrahydrofolate + dUMP = dihydrofolate + dTMP. |
- General function:
- Involved in acid phosphatase activity
- Specific function:
- Dephosphorylates several organic phosphomonoesters and catalyzes the transfer of low-energy phosphate groups from phosphomonoesters to hydroxyl groups of various organic compounds. Preferentially acts on aryl phosphoesters. Might function as a broad-spectrum dephosphorylating enzyme able to scavenge both 3'- and 5'-nucleotides and also additional organic phosphomonoesters
- Gene Name:
- aphA
- Locus Tag:
- PA1409
- Molecular weight:
- 38 kDa
Reactions
| A phosphate monoester + H(2)O = an alcohol + phosphate. |
- General function:
- Involved in nucleoside-triphosphate diphosphatase activity
- Specific function:
- Specific function unknown
- Gene Name:
- mazG
- Locus Tag:
- PA0935
- Molecular weight:
- 31.2 kDa
Reactions
| ATP + H(2)O = AMP + diphosphate. |