ADP (PAMDB110479)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB110479 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | An organophosphate oxoanion that is the trianion of adenosine 5'-diphosphate arising from deprotonation of all three OH groups of the diphosphate; major species present at pH 7.3. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
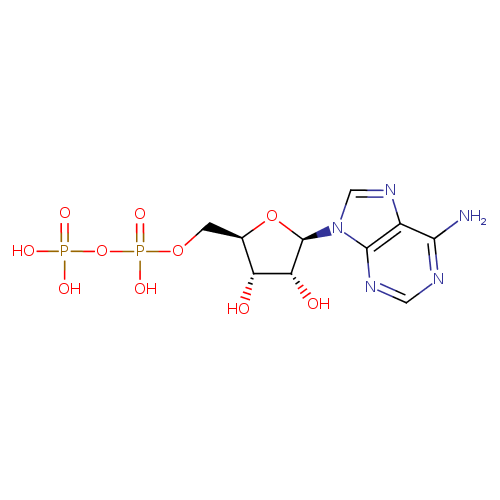
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H12N5O10P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 424.18 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 427.0294147485 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | XTWYTFMLZFPYCI-KQYNXXCUSA-K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H15N5O10P2/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(24-10)1-23-27(21,22)25-26(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H2,11,12,13)(H2,18,19,20)/p-3/t4-,6-,7-,10-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 58-64-0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | adenosine 5'-diphosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | adenosine-diphosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C(C3(C(C(C(N2(C1(=C(C(=NC=N1)N)N=C2)))O3)O)O))OP(OP(=O)([O-])[O-])([O-])=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of chemical entities known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Chemical entities | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside diphosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 2'-deoxyribose + ATP → DEOXY-RIBOSE-5P + ADP + Hydrogen ion ATP + dAMP → ADP + dADP coenzyme A + acetate + ATP → phosphate + acetyl-CoA + ADP pseudouridine + ATP → Hydrogen ion + pseudouridine 5'-phosphate + ADP ATP + acetyl-CoA + Water → malonyl-CoA + ADP + phosphate + Hydrogen ion dGMP + ATP → dGDP + ADP ATP + DNA-N + Water → ADP + phosphate ATP + protoporphyrin IX + MG+2 + Water → Hydrogen ion + Mg-protoporphyrin + phosphate + ADP ATP + dIDP → ADP + dITP beta-Alanine + (R)-pantoate + ATP → Hydrogen ion + (R)-pantothenate + phosphate + ADP Deoxyadenosine + ATP → Hydrogen ion + dAMP + ADP dADP + Water → ADP UDP-N-acetylmuramoyl-L-alanyl-γ-D-glutamyl-L-lysine + D-Alanyl-D-alanine + ATP → Hydrogen ion + UDP-N-acetyl-α-D-muramoyl-L-alanyl-γ-D-glutamyl-L-lysyl-D-alanyl-D-alanine + phosphate + ADP 2-dehydro-3-deoxy-D-gluconate + ATP → Hydrogen ion + 2-dehydro-3-deoxy-D-gluconate 6-phosphate + ADP ATP + dCMP → ADP + dCDP Deoxyguanosine + ATP → Hydrogen ion + dGMP + ADP Protein-L-serine-or-L-threonine + ATP → Protein-Ser-or-Thr-phosphate + ADP + Hydrogen ion ATP + 4-Amino-5-hydroxymethyl-2-methylpyrimidine → Hydrogen ion + ADP + 4-Amino-2-methyl-5-phosphomethylpyrimidine Glucopyranose + ATP → D-glucopyranose-6-phosphate + ADP + Hydrogen ion chlorophyllide a + ADP + phosphate → protochlorophyllide a + ATP + Water METHYLENE-THF-GLU-N + L-glutamate + ATP → METHYLENE-THF-GLU-N + ADP + phosphate More...alpha-D-Glucose + ATP → Hydrogen ion + α-D-glucose 6-phosphate + ADP 5-oxoproline + Water + ATP → Hydrogen ion + L-glutamate + phosphate + ADP Hydrogen ion + ATP + Water → Hydrogen ion + ADP + phosphate alpha-N-Peptidyl-LGlutamate + L-glutamate + ATP → CPD0-2471 + ADP + phosphate + Hydrogen ion thiamin + ATP → Hydrogen ion + thiamin phosphate + ADP α-D-Kdo-(2→4)-α-D-Kdo-(2→6)-lipid A + ADP-L-glycero-beta-D-manno-heptose → heptosyl-Kdo2-lipid A + ADP + Hydrogen ion ALGINATE + Water + ATP → ALGINATE + phosphate + ADP + Hydrogen ion dADP + ATP → dATP + ADP LysW-L-glutamate + ATP → LysW-L-glutamate-5-phosphate + ADP dGDP + ATP → dGTP + ADP α-Kdo-(2->4)-α-Kdo-(2->6)-lipid IVA + ADP-L-glycero-beta-D-manno-heptose → Hydrogen ion + heptosyl-Kdo2-lipid IVA + ADP D-ALPHABETA-D-HEPTOSE-7-PHOSPHATE + ATP → Hydrogen ion + D-glycero-β-D-manno-heptose 1,7-bisphosphate + ADP 1,6-anhydro-N-acetyl-β-muramate + ATP + Water → Hydrogen ion + N-acetyl-β-muramate 6-phosphate + ADP ATP → ADP cob(II)yrinate c-monoamide + L-Glutamine + ATP + Water → cob(II)yrinate a,c-diamide + L-glutamate + ADP + phosphate + Hydrogen ion ATP + acetyl-CoA + Hydrogen carbonate → Hydrogen ion + malonyl-CoA + phosphate + ADP cobyrinate + L-Glutamine + ATP + Water → cob(II)yrinate c-monoamide + L-glutamate + ADP + phosphate + Hydrogen ion ATP → ADP + phosphate Water + ATP → Hydrogen ion + phosphate + ADP cobyrinate + L-Glutamine + ATP + Water → cob(II)yrinate a,c-diamide + L-glutamate + ADP + phosphate + Hydrogen ion Nucleoside-Diphosphates + ATP → Nucleoside-Triphosphates + ADP L-Glutamine + ATP + Water → L-glutamate + phosphate + ADP + Hydrogen ion Xenobiotic + Water + ATP → Xenobiotic + phosphate + ADP GDP + ADP → GTP + AMP galactosyl-(glucosyl)3-(heptosyl)3-Kdo2-lipid A-bisphosphate + ADP-L-glycero-beta-D-manno-heptose → Hydrogen ion + lipid A-core + ADP UDP-N-acetyl-α-D-muramate + L-alanyl-γ-D-glutamyl-meso-diaminopimelate + ATP → Hydrogen ion + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate + ADP + phosphate L-glutamate + LysW-C-Terminal-L-Glutamate + ATP → LysW-L-glutamate + ADP + phosphate + Hydrogen ion cis-geranyl-CoA + Hydrogen carbonate + ATP → 3-(4-methylpent-3-en-1-yl)-pent-2-enedioyl-CoA + phosphate + ADP + Hydrogen ion NAD+ + ATP → Hydrogen ion + NADP+ + ADP glucosyl-(heptosyl)3-Kdo2-lipid A-phosphate + ATP → Hydrogen ion + glucosyl-(heptosyl)3-Kdo2-lipid A-bisphosphate + ADP ATP + Water + K+ → ADP + phosphate + K+ + Hydrogen ion glucosyl-(heptosyl)2-Kdo2-lipid A-phosphate + ADP-L-glycero-beta-D-manno-heptose → Hydrogen ion + glucosyl-(heptosyl)3-Kdo2-lipid A-phosphate + ADP β-D-fructofuranose 6-phosphate + ADP → Hydrogen ion + fructose 1,6-bisphosphate + AMP glucosyl-(heptosyl)2-Kdo2-lipid A + ATP → Hydrogen ion + glucosyl-(heptosyl)2-Kdo2-lipid A-phosphate + ADP ATP + IDP → ITP + ADP L-1-PHOSPHATIDYL-GLYCEROL + ATP → L-1-PHOSPHATIDYL-GLYCEROL-P + ADP + Hydrogen ion heptosyl-Kdo2-lipid A + ADP-L-glycero-beta-D-manno-heptose → Hydrogen ion + (heptosyl)2-Kdo2-lipid A + ADP ATP + 4-methyl-5-(β-hydroxyethyl)thiazole → Hydrogen ion + ADP + 4-methyl-5-(2-phosphonooxyethyl)thiazole | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Yamagata, Yukio. Prebiotic formation of ADP and ATP from AMP, calcium phosphates and cyanate in aqueous solution. Origins of Life and Evolution of the Biosphere (1999), 29(5), 511-520. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||