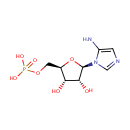| Identification |
| Name: |
Phosphoribosylaminoimidazole carboxylase ATPase subunit |
| Synonyms: |
|
| Gene Name: |
purK |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in phosphoribosylaminoimidazole carboxylase activity |
| Specific Function: |
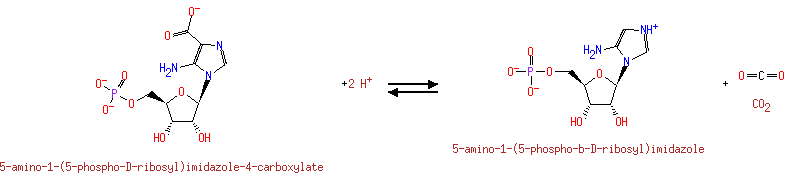
Possesses an ATPase activity that is dependent on the presence of AIR (aminoimidazole ribonucleotide). The association of purK and purE produces an enzyme complex capable of converting AIR to CAIR efficiently under physiological condition |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
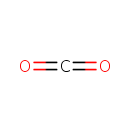
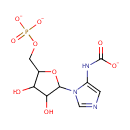
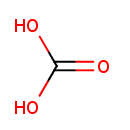
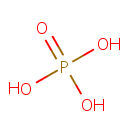
 | + | 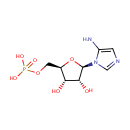 | + | 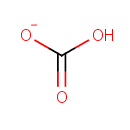 | ↔ |  | + | 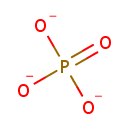 | + | 5-Carboxyamino-1-(5-phospho-D-ribosyl)imidazole | + | 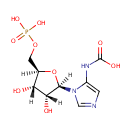 |
| | | |
|
| SMPDB Reactions: |
|
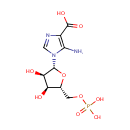
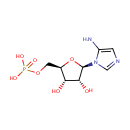 | + |  | + |  | → | N5-Carboxyaminoimidazole ribonucleotide | + | Adenosine diphosphate | + |  | + | 2  | + | 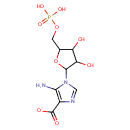 | + |  |
| | | |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | carbon-carbon lyase activity | | carboxy-lyase activity | | catalytic activity | | lyase activity | | nucleoside binding | | phosphoribosylaminoimidazole carboxylase activity | | purine nucleoside binding | | Process |
|---|
| 'de novo' IMP biosynthetic process | | cellular nitrogen compound metabolic process | | IMP biosynthetic process | | metabolic process | | nitrogen compound metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleoside phosphate metabolic process | | nucleotide metabolic process | | purine nucleoside monophosphate biosynthetic process | | purine nucleotide biosynthetic process | | purine nucleotide metabolic process | | purine ribonucleoside monophosphate biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA5425 |
| Strand: |
- |
| Entrez Gene ID: |
878440 |
| Accession: |
NP_254112.1 |
| GI: |
15600618 |
| Sequence start: |
6105159 |
| Sequence End: |
6106241 |
| Sequence Length: |
1082 |
| Gene Sequence: |
>PA5425
ATGAAAATCGGTGTCATCGGTGGCGGCCAGCTGGGCCGCATGCTGGCCCTGGCGGGAACCCCGCTGGGCATGAACTTCGCTTTCCTCGACCCGGCGCCGGACGCCTGCGCGGCCTCCCTGGGCGAGCACATCCGCGCCGACTACGGCGACCAGGAGCACCTGCGGCAACTGGCCGACGAGGTCGACCTGGTAACCTTCGAGTTCGAGAGCGTGCCGGCGGAAACCGTGGCCTTCCTCTCCCAGTTCGTGCCGGTCTACCCGAACGCCGAGTCGCTGCGCATCGCCCGCGACCGCTGGTTCGAGAAGTCGATGTTCAAGGACCTTGGCATTCCCACTCCGGACTTCGCCGACGTCCAGTCCCAGGCCGACCTCGATGCCGCCGCAGCCGCCATCGGCCTGCCGGCGGTGCTCAAGACCCGCACCCTGGGCTACGACGGCAAGGGCCAGAAGGTCCTGCGCCAGCCGGCCGACGTGCAGGGCGCGTTCGCCGAACTGGGCAGCGTGCCGTGCATCCTCGAGGGCTTCGTGCCGTTCACCGGGGAAGTCTCGCTGGTGGCGGTGCGCGCTCGCGATGGCGAGACGCGTTTCTACCCGCTGGTGCACAACACCCACGACAGCGGCATCCTCAAGCTCTCCGTGGCCAGCAGCGGGCATCCGTTGCAGGCGCTGGCCGAGGACTACGTCGGCCGCGTGCTGGCCCGGCTCGACTACGTCGGCGTGCTGGCCTTCGAGTTCTTCGAGGTGGACGGCGGCCTGAAGGCCAACGAGATCGCCCCGCGCGTGCACAACTCCGGGCACTGGACCATCGAAGGCGCCGAGTGCAGCCAGTTCGAGAACCACTTGCGCGCCGTCGCCGGCCTGCCGCTGGGCTCGACCGCCAAGGTCGGCGAGAGCGCGATGCTCAATTTCATCGGCGCGGTCCCCCCGGTGGCTCAGGTGGTCGCCGTCGCCGACTGCCACCTGCATCACTACGGCAAGGCCTTCAAGAACGGCCGCAAGGTCGGCCACGCCACCCTGCGTTGTGCCGACCGGGCGACGCTGCAGGCGCGCATCGCCGAGGTCGAGGCGCTGATCGAGGCGTGA |
| Protein Properties |
| Protein Residues: |
360 |
| Protein Molecular Weight: |
38.5 kDa |
| Protein Theoretical pI: |
5.38 |
| Hydropathicity (GRAVY score): |
0.082 |
| Charge at pH 7 (predicted): |
-10.26 |
| Protein Sequence: |
>PA5425
MKIGVIGGGQLGRMLALAGTPLGMNFAFLDPAPDACAASLGEHIRADYGDQEHLRQLADEVDLVTFEFESVPAETVAFLSQFVPVYPNAESLRIARDRWFEKSMFKDLGIPTPDFADVQSQADLDAAAAAIGLPAVLKTRTLGYDGKGQKVLRQPADVQGAFAELGSVPCILEGFVPFTGEVSLVAVRARDGETRFYPLVHNTHDSGILKLSVASSGHPLQALAEDYVGRVLARLDYVGVLAFEFFEVDGGLKANEIAPRVHNSGHWTIEGAECSQFENHLRAVAGLPLGSTAKVGESAMLNFIGAVPPVAQVVAVADCHLHHYGKAFKNGRKVGHATLRCADRATLQARIAEVEALIEA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5425 and its homologs
|