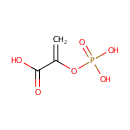
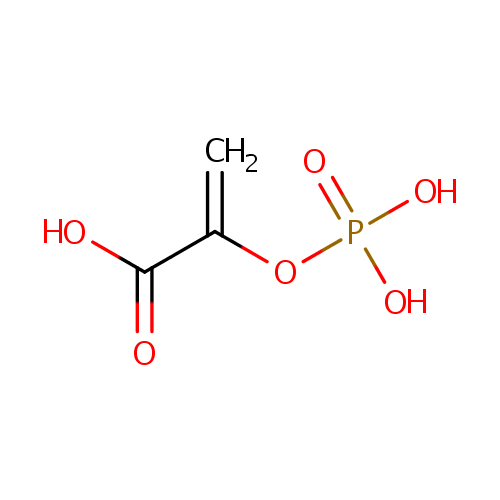
Phosphoenolpyruvic acid (PAMDB000111)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000111 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Phosphoenolpyruvic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Phosphoenolpyruvate (PEP) plays a key role in many metabolic reactions. It has a high energy phosphate bond, and is involved in glycolysis and gluconeogenesis. In glycolysis, PEP is formed by the action of the enzyme enolase on 2-phosphoglycerate. Metabolism of PEP to pyruvate by pyruvate kinase (PK) generates 1 molecule of adenosine triphosphate (ATP) via substrate-level phosphorylation. ATP is one of the major currencies of chemical energy within cells. In gluconeogenesis, PEP is formed from the decarboxylation of oxaloacetate and hydrolysis of 1 guanosine triphosphate molecule. This reaction is catalyzed by the enzyme phosphoenolpyruvate carboxykinase (PEPCK). This reaction is a rate-limiting step in gluconeogenesis. (wikipedia) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H5O6P | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 168.042 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 167.982374404 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | DTBNBXWJWCWCIK-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H5O6P/c1-2(3(4)5)9-10(6,7)8/h1H2,(H,4,5)(H2,6,7,8) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 138-08-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-(phosphonooxy)prop-2-enoic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | phosphoenolpyruvic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(=O)C(=C)OP(O)(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as phosphate esters. These are organic compounds containing phosphoric acid ester functional group, with the general structure R1P(=O)(R2)OR3. R1,R2 = O,N, or halogen atom; R3 = organyl group. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organophosphorus compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organic phosphoric acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Phosphate esters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Phosphate esters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Phosphoenolpyruvic acid + N-Acetyl-D-glucosamine > N-Acetyl-D-Glucosamine 6-Phosphate + Pyruvic acid Phosphoenolpyruvic acid + D-Glucose > Glucose 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + 2(alpha-D-Mannosyl)-D-glycerate > 2(alpha-D-Mannosyl-6-phosphate)-D-glycerate + Pyruvic acid Dihydroxyacetone + Phosphoenolpyruvic acid > Dihydroxyacetone phosphate + Pyruvic acid Phosphoenolpyruvic acid + D-Mannose > Mannose 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + D-Fructose > Fructose 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + N-Acetylmannosamine > N-Acetyl-D-mannosamine 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Glucosamine > Glucosamine 6-phosphate + Pyruvic acid ADP + Hydrogen ion + Phosphoenolpyruvic acid <> Adenosine triphosphate + Pyruvic acid Phosphoenolpyruvic acid + Galactitol > Galactitol 1-phosphate + Pyruvic acid Phosphoenolpyruvic acid + D-Fructose > Fructose 1-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Sorbitol > Pyruvic acid + Sorbitol-6-phosphate Phosphoenolpyruvic acid + Ascorbic acid > L-Ascorbate 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + D-Maltose > Maltose 6'-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Trehalose > Pyruvic acid + Trehalose 6-phosphate Phosphoenolpyruvic acid + Sucrose > Pyruvic acid + Sucrose-6-phosphate Phosphoenolpyruvic acid + N-Acetyl-D-muramoate > N-Acetylmuramic acid 6-phosphate + Pyruvic acid D-Erythrose 4-phosphate + Water + Phosphoenolpyruvic acid <> 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate + Phosphate Phosphoenolpyruvic acid + Mannitol > Sorbitol-6-phosphate + Pyruvic acid D-Arabinose 5-phosphate + Water + Phosphoenolpyruvic acid <> 3-Deoxy-D-manno-octulosonate 8-phosphate + Phosphate Adenosine triphosphate + Water + Pyruvic acid <> Adenosine monophosphate +2 Hydrogen ion + Phosphoenolpyruvic acid + Phosphate Phosphoenolpyruvic acid + Chitobiose > Diacetylchitobiose-6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Uridine diphosphate-N-acetylglucosamine <> Phosphate + UDP-N-Acetyl-3-(1-carboxyvinyl)-D-glucosamine More...Adenosine triphosphate + Oxalacetic acid <> ADP + Carbon dioxide + Phosphoenolpyruvic acid Carbon dioxide + Water + Phosphoenolpyruvic acid <> Hydrogen ion + Oxalacetic acid + Phosphate + Hydrogen carbonate Adenosine triphosphate + Pyruvic acid + Water <> Adenosine monophosphate + Phosphoenolpyruvic acid + Phosphate Adenosine triphosphate + Pyruvic acid <> ADP + Phosphoenolpyruvic acid Phosphate + Oxalacetic acid <> Water + Phosphoenolpyruvic acid + Carbon dioxide Guanosine triphosphate + Pyruvic acid <> Guanosine diphosphate + Phosphoenolpyruvic acid dATP + Pyruvic acid <> dADP + Phosphoenolpyruvic acid dGTP + Pyruvic acid <> dGDP + Phosphoenolpyruvic acid Nucleoside triphosphate + Pyruvic acid <> NDP + Phosphoenolpyruvic acid Phosphoenolpyruvic acid + b-D-Glucose > Glucose 6-phosphate + Pyruvic acid Shikimate 3-phosphate + Phosphoenolpyruvic acid <> 5-O-(1-Carboxyvinyl)-3-phosphoshikimate + Phosphate 2-Phospho-D-glyceric acid <> Phosphoenolpyruvic acid + Water Phosphate + Oxalacetic acid <> Phosphoenolpyruvic acid + Hydrogen carbonate Phosphoenolpyruvic acid + <i>N</i>-acetylmuramate > N-Acetylmuramic acid 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Salicin > Salicin 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Cellobiose > Cellobiose-6-phosphate + Pyruvic acid D-fructose + Phosphoenolpyruvic acid > Fructose 1-phosphate + Pyruvic acid D-fructose + Phosphoenolpyruvic acid > Fructose 6-phosphate + Pyruvic acid Phosphoenolpyruvic acid + Shikimate 3-phosphate > Inorganic phosphate + 5-O-(1-Carboxyvinyl)-3-phosphoshikimate Phosphoenolpyruvic acid + D-Erythrose 4-phosphate + Water > 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate + Inorganic phosphate Inorganic phosphate + Oxalacetic acid > Water + Phosphoenolpyruvic acid + Carbonic acid Phosphoenolpyruvic acid + protein L-histidine > Pyruvic acid + protein N(pi)-phospho-L-histidine Phosphoenolpyruvic acid + D-Arabinose 5-phosphate + Water > 3-Deoxy-D-manno-octulosonate 8-phosphate + Inorganic phosphate Phosphoenolpyruvic acid + Uridine diphosphate-N-acetylglucosamine > Inorganic phosphate + UDP-N-Acetyl-3-(1-carboxyvinyl)-D-glucosamine Adenosine triphosphate + Pyruvic acid + Water > Adenosine monophosphate + Phosphoenolpyruvic acid + Inorganic phosphate Phosphoenolpyruvic acid + Protein histidine <> Pyruvic acid + Protein N(pi)-phospho-L-histidine Phosphoenolpyruvic acid > Water + 2-Phosphoglyceric acid + 2-Phosphoglyceric acid Phosphoenolpyruvic acid + Adenosine diphosphate + Hydrogen ion + ADP > Adenosine triphosphate + Pyruvic acid D-Erythrose 4-phosphate + Water + Phosphoenolpyruvic acid > Phosphate + 3-deoxy-D-arabino-heptulosonate-7-phosphate shikimate 3-phosphate + Phosphoenolpyruvic acid + Shikimate 3-phosphate > Phosphate + 5-enolpyruvyl-shikimate 3-phosphate Oxalacetic acid + Adenosine triphosphate > Adenosine diphosphate + Carbon dioxide + Phosphoenolpyruvic acid + ADP D-Arabinose 5-phosphate + Phosphoenolpyruvic acid + Water > Phosphate + 3-deoxy-D-manno-octulosonate 8-phosphate + 3-Deoxy-D-manno-octulosonate 8-phosphate Uridine diphosphate-N-acetylglucosamine + Phosphoenolpyruvic acid > Phosphate + UDP-N-acetyl-α-D-glucosamine-enolpyruvate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Simon, Ethan S.; Grabowski, Sven; Whitesides, George M. Preparation of phosphoenolpyruvate from D-(-)-3-phosphoglyceric acid for use in regeneration of ATP. Journal of the American Chemical Society (1989), 111(24), 8920-1. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in phosphoenolpyruvate carboxylase activity
- Specific function:
- Through the carboxylation of phosphoenolpyruvate (PEP) it forms oxaloacetate, a four-carbon dicarboxylic acid source for the tricarboxylic acid cycle
- Gene Name:
- ppc
- Locus Tag:
- PA3687
- Molecular weight:
- 97.8 kDa
Reactions
| Phosphate + oxaloacetate = H(2)O + phosphoenolpyruvate + HCO(3)(-). |
- General function:
- Involved in catalytic activity
- Specific function:
- Stereospecific condensation of phosphoenolpyruvate (PEP) and D-erythrose-4-phosphate (E4P) giving rise to 3-deoxy-D- arabino-heptulosonate-7-phosphate (DAHP)
- Gene Name:
- aroF
- Locus Tag:
- PA1750
- Molecular weight:
- 39.1 kDa
Reactions
| Phosphoenolpyruvate + D-erythrose 4-phosphate + H(2)O = 3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate + phosphate. |
- General function:
- Involved in protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in N-acetylglucosamine transport
- Gene Name:
- nagE
- Locus Tag:
- PA3761
- Molecular weight:
- 60.6 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the reversible conversion of 2- phosphoglycerate into phosphoenolpyruvate. It is essential for the degradation of carbohydrates via glycolysis. It is also a component of the RNA degradosome, a multi-enzyme complex involved in RNA processing and messenger RNA degradation. Its interaction with RNase E is important for the turnover of mRNA, in particular on transcripts encoding enzymes of energy-generating metabolic routes. Its presence in the degradosome is required for the response to excess phosphosugar. May play a regulatory role in the degradation of specific RNAs, such as ptsG mRNA, therefore linking cellular metabolic status with post-translational gene regulation
- Gene Name:
- eno
- Locus Tag:
- PA3635
- Molecular weight:
- 45.2 kDa
Reactions
| 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O. |
- General function:
- Involved in catalytic activity
- Specific function:
- Synthesis of KDO 8-P which is required for lipid A maturation and cellular growth
- Gene Name:
- kdsA
- Locus Tag:
- PA3636
- Molecular weight:
- 31.1 kDa
Reactions
| Phosphoenolpyruvate + D-arabinose 5-phosphate + H(2)O = 2-dehydro-3-deoxy-D-octonate 8-phosphate + phosphate. |
- General function:
- Involved in transferase activity, transferring alkyl or aryl (other than methyl) groups
- Specific function:
- Cell wall formation. Adds enolpyruvyl to UDP-N- acetylglucosamine. Target for the antibiotic phosphomycin
- Gene Name:
- murA
- Locus Tag:
- PA4450
- Molecular weight:
- 44.6 kDa
Reactions
| Phosphoenolpyruvate + UDP-N-acetyl-D-glucosamine = phosphate + UDP-N-acetyl-3-O-(1-carboxyvinyl)-D-glucosamine. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- ATP + pyruvate = ADP + phosphoenolpyruvate
- Gene Name:
- pykF
- Locus Tag:
- PA1498
- Molecular weight:
- 51.5 kDa
Reactions
| ATP + pyruvate = ADP + phosphoenolpyruvate. |
- General function:
- Involved in sugar:hydrogen symporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in fructose transport
- Gene Name:
- fruA
- Locus Tag:
- PA3560
- Molecular weight:
- 59 kDa
Reactions
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- ATP + pyruvate = ADP + phosphoenolpyruvate
- Gene Name:
- pykA
- Locus Tag:
- PA4329
- Molecular weight:
- 52.3 kDa
Reactions
| ATP + pyruvate = ADP + phosphoenolpyruvate. |
- General function:
- Involved in phosphoenolpyruvate carboxykinase (ATP) activity
- Specific function:
- ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO(2)
- Gene Name:
- pckA
- Locus Tag:
- PA5192
- Molecular weight:
- 55.7 kDa
Reactions
| ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the phosphorylation of pyruvate to phosphoenolpyruvate
- Gene Name:
- ppsA
- Locus Tag:
- PA1770
- Molecular weight:
- 85.8 kDa
Reactions
| ATP + pyruvate + H(2)O = AMP + phosphoenolpyruvate + phosphate. |
- General function:
- Involved in protein binding
- Specific function:
- Component of the phosphoenolpyruvate-dependent nitrogen- metabolic phosphotransferase system (nitrogen-metabolic PTS), that seems to be involved in regulating nitrogen metabolism. Enzyme I- Ntr transfers the phosphoryl group from phosphoenolpyruvate (PEP) to the phosphoryl carrier protein (NPr). Could function in the transcriptional regulation of sigma-54 dependent operons in conjunction with the NPr (ptsO) and EIIA-Ntr (ptsN) proteins
- Gene Name:
- ptsP
- Locus Tag:
- PA0337
- Molecular weight:
- 83.6 kDa
Reactions
| Phosphoenolpyruvate + protein L-histidine = pyruvate + protein N(pi)-phospho-L-histidine. |
- General function:
- Involved in sugar:hydrogen symporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatB
- Locus Tag:
- PA4484
- Molecular weight:
- 53.1 kDa
Reactions
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in transporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatA
- Locus Tag:
- PA4483
- Molecular weight:
- 51.9 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatC
- Locus Tag:
- PA4482
- Molecular weight:
- 10.5 kDa
Transporters
- General function:
- Involved in protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in N-acetylglucosamine transport
- Gene Name:
- nagE
- Locus Tag:
- PA3761
- Molecular weight:
- 60.6 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in sugar:hydrogen symporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in fructose transport
- Gene Name:
- fruA
- Locus Tag:
- PA3560
- Molecular weight:
- 59 kDa
Reactions
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatC
- Locus Tag:
- PA4482
- Molecular weight:
- 10.5 kDa