| Identification |
| Name: |
UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
| Synonyms: |
- Enoylpyruvate transferase
- UDP-N-acetylglucosamine enolpyruvyl transferase
- EPT
|
| Gene Name: |
murA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in transferase activity, transferring alkyl or aryl (other than methyl) groups |
| Specific Function: |
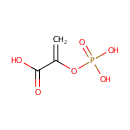
Cell wall formation. Adds enolpyruvyl to UDP-N- acetylglucosamine. Target for the antibiotic phosphomycin |
| Cellular Location: |
Cytoplasm (Probable) |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
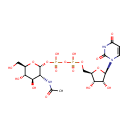
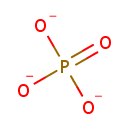
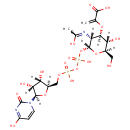
| PAMDB001776 | Inorganic phosphate | MetaboCard | | PAMDB000383 | Phosphate | MetaboCard | | PAMDB000111 | Phosphoenolpyruvic acid | MetaboCard | | PAMDB000647 | UDP-N-Acetyl-3-(1-carboxyvinyl)-D-glucosamine | MetaboCard | | PAMDB006354 | UDP-N-acetyl-α-D-glucosamine-enolpyruvate | MetaboCard | | PAMDB000120 | Uridine diphosphate-N-acetylglucosamine | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | transferase activity | | transferase activity, transferring alkyl or aryl (other than methyl) groups | | Process |
|---|
| alcohol metabolic process | | amino sugar metabolic process | | metabolic process | | monosaccharide metabolic process | | small molecule metabolic process | | UDP-N-acetylgalactosamine biosynthetic process | | UDP-N-acetylgalactosamine metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4450 |
| Strand: |
- |
| Entrez Gene ID: |
880969 |
| Accession: |
NP_253140.1 |
| GI: |
15599646 |
| Sequence start: |
4984205 |
| Sequence End: |
4985470 |
| Sequence Length: |
1265 |
| Gene Sequence: |
>PA4450
ATGGATAAACTGATTATTACCGGCGGTAACCGCCTCGATGGCGAAATCCGCATTTCCGGCGCGAAGAACTCGGCGCTGCCGATCCTGGCCGCGACCCTGCTGGCCGATACTCCGGTCACCGTCTGCAACCTGCCGCACCTGCACGACATTACCACCATGATCGAACTGTTCGGCCGCATGGGCGTGCAGCCGATCATCGACGAGAAGCTCAACGTCGAAGTCGATGCCAGCAGCATCAAAACCCTGGTCGCGCCGTACGAACTGGTGAAGACCATGCGTGCCTCGATCCTGGTGCTGGGCCCGATGCTGGCGCGCTTCGGCGAGGCCGAAGTGGCCCTGCCGGGCGGTTGCGCGATCGGTTCGCGTCCGGTCGACCTGCATATCCGCGGTCTCGAGGCCATGGGCGCGCAGATCGAGGTCGAAGGCGGCTACATCAAGGCCAAGGCGCCGGCCGGCGGCCTGCGTGGCGGTCACTTCTTCTTCGATACCGTCAGCGTGACCGGCACCGAGAACCTGATGATGGCCGCCGCGCTGGCCAACGGCCGTACCGTGCTGCAGAACGCCGCTCGCGAGCCGGAGGTGGTCGACCTGGCCAACTGCCTGAACGCCATGGGCGCCAACGTCCAGGGCGCTGGCTCCGATACCATCGTCATCGAAGGCGTGAAGCGCCTCGGCGGTGCTCGCTACGACGTACTGCCCGACCGTATCGAGACCGGCACCTACCTGGTGGCCGCGGCCGCGACCGGTGGCCGGGTGAAGCTGAAGGATACCGACCCGACCATCCTCGAGGCGGTCCTGCAGAAGCTGGAAGAGGCCGGTGCCCACATCAGCACCGGCAGCAACTGGATCGAGCTGGACATGAAGGGCAACCGGCCGAAGGCGGTCAACGTGCGTACCGCGCCGTACCCGGCGTTCCCCACCGACATGCAGGCCCAGTTCATCTCCATGAACGCGGTAGCCGAAGGCACCGGCGCGGTCATCGAGACGGTCTTCGAGAACCGCTTCATGCATGTTTACGAAATGAACCGCATGGGCGCGCAAATCCTCGTCGAAGGCAACACCGCCATCGTCACCGGCGTACCCAAGCTCAAGGGCGCTCCGGTCATGGCGACCGACCTGCGCGCATCCGCGAGCCTGGTGATCGCCGGCCTGGTGGCCGAAGGCGACACCCTGATCGATCGCATCTACCACATCGACCGTGGCTACGAGTGCATCGAAGAGAAACTCCAGCTGCTCGGCGCCAAGATCCGCCGCGTACCGGGCTAG |
| Protein Properties |
| Protein Residues: |
421 |
| Protein Molecular Weight: |
44.6 kDa |
| Protein Theoretical pI: |
5.49 |
| Hydropathicity (GRAVY score): |
0.152 |
| Charge at pH 7 (predicted): |
-6.42 |
| Protein Sequence: |
>PA4450
MDKLIITGGNRLDGEIRISGAKNSALPILAATLLADTPVTVCNLPHLHDITTMIELFGRMGVQPIIDEKLNVEVDASSIKTLVAPYELVKTMRASILVLGPMLARFGEAEVALPGGCAIGSRPVDLHIRGLEAMGAQIEVEGGYIKAKAPAGGLRGGHFFFDTVSVTGTENLMMAAALANGRTVLQNAAREPEVVDLANCLNAMGANVQGAGSDTIVIEGVKRLGGARYDVLPDRIETGTYLVAAAATGGRVKLKDTDPTILEAVLQKLEEAGAHISTGSNWIELDMKGNRPKAVNVRTAPYPAFPTDMQAQFISMNAVAEGTGAVIETVFENRFMHVYEMNRMGAQILVEGNTAIVTGVPKLKGAPVMATDLRASASLVIAGLVAEGDTLIDRIYHIDRGYECIEEKLQLLGAKIRRVPG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4450 and its homologs
|