| Identification |
| Name: |
Phosphoenolpyruvate-protein phosphotransferase ptsP |
| Synonyms: |
- Enzyme I-Ntr
- Phosphotransferase system, enzyme I
|
| Gene Name: |
ptsP |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in protein binding |
| Specific Function: |
Component of the phosphoenolpyruvate-dependent nitrogen- metabolic phosphotransferase system (nitrogen-metabolic PTS), that seems to be involved in regulating nitrogen metabolism. Enzyme I- Ntr transfers the phosphoryl group from phosphoenolpyruvate (PEP) to the phosphoryl carrier protein (NPr). Could function in the transcriptional regulation of sigma-54 dependent operons in conjunction with the NPr (ptsO) and EIIA-Ntr (ptsN) proteins |
| Cellular Location: |
Cytoplasm (Probable) |
| KEGG Pathways: |
- Phosphotransferase system (PTS) pae02060
|
| KEGG Reactions: |
|
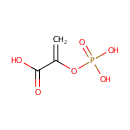 | + | Protein histidine | ↔ | 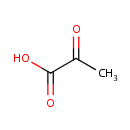 | + | Protein N(pi)-phospho-L-histidine |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
 | + | protein L-histidine | → |  | + | protein N(pi)-phospho-L-histidine |
| | |
 | + | Protein histidine | ↔ |  | + | Protein N(pi)-phospho-L-histidine |
| | |
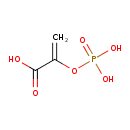 | + | PTS-I-Histidines | → | 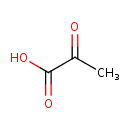 | + | PTS-I-pi-phospho-L-histidines |
| |
|
| Complex Reactions: |
|
 | + | protein L-histidine | → |  | + | protein N(pi)-phospho-L-histidine |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| catalytic activity | | cation transmembrane transporter activity | | cation:sugar symporter activity | | ion transmembrane transporter activity | | phosphoenolpyruvate-protein phosphotransferase activity | | phosphotransferase activity, nitrogenous group as acceptor | | solute:cation symporter activity | | substrate-specific transmembrane transporter activity | | sugar:hydrogen symporter activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | transmembrane transporter activity | | transporter activity | | Process |
|---|
| carbohydrate transport | | cellular metabolic process | | establishment of localization | | metabolic process | | phosphate metabolic process | | phosphoenolpyruvate-dependent sugar phosphotransferase system | | phosphorus metabolic process | | phosphorylation | | transport |
|
| Gene Properties |
| Locus tag: |
PA0337 |
| Strand: |
+ |
| Entrez Gene ID: |
882232 |
| Accession: |
NP_249028.1 |
| GI: |
15595534 |
| Sequence start: |
378598 |
| Sequence End: |
380877 |
| Sequence Length: |
2279 |
| Gene Sequence: |
>PA0337
ATGCTCAACACGCTGCGCAAGATCGTCCAGGAAGTGAACTCCGCCAAGGATCTGAAGGCGGCGCTGGGCATCATCGTGCAGCGGGTCAAGGAAGCCATGGGTACCCAGGTCTGCTCGGTGTACCTGCTCGACACCGAGACCCAGCGTTTCGTCCTGATGGCCACCGAAGGCCTCAACAAGCGTTCCATCGGCAAGGTCAGCATGGCTCCCAGCGAAGGCCTGGTCGGCCTGGTCGGCACCCGCGAGGAGCCGCTCAACCTGGAGAACGCCGCCGCCCACCCGCGCTACCGCTATTTCGCCGAGACCGGCGAGGAGCGCTACGCGTCGTTCCTCGGCGCGCCGATCATCCACCATCGGCGGGTGATGGGGGTGCTGGTGGTGCAGCAGAAGGAGCGCCGCCAGTTCGACGAAGGCGAGGAAGCCTTCCTCGTCACCATGAGCGCCCAGCTCGCCGGGGTCATCGCGCATGCCGAGGCGACCGGTTCGATCCGCGGCCTGGGCAAGCTCGGCAAGGGCATCCAGGAAGCCAAGTTCGTCGGCGTGCCCGGCGCCCCCGGGGTCGGGGTGGGCAAGGCGGTGGTGGTGTTGCCGCCGGCCGACCTGGAAGTGGTGCCGGACAAGCAGGTCGACGACATCGACGCCGAGATCGCCCTGTTCAAGCAGGCCCTGGAGGGCGTTCGCGCCGACATGCGCGCGCTGTCGAGCAAGCTCGCCAGCCAGCTGCGCAAGGAAGAACGCGCGCTGTTCGACGTCTACCTGATGATGCTCGACGATGCCTCCATCGGCAACGAGGTCAAGCGCATCATCCGTACCGGCCAGTGGGCCCAGGGCGCCCTGCGCCAGGTGGTGATGGAGCACGTGCAGCGCTTCGAGCTGATGGACGACGCCTATCTCCGCGAGCGCGCCTCCGACGTCAAGGACATAGGTCGCCGCCTGCTCGCCTACCTCCAGGAAGAGCGCAAGCAGAACCTGACCTACCCGGAACAGACCATCATCGTCAGCGAGGAGCTGTCGCCGGCGATGCTCGGCGAGGTGCCGGAAGGGCGCCTGGTCGGCCTGGTCTCGGTGCTCGGCTCGGGCAACTCGCACGTGGCGATCCTCGCCCGTGCCATGGGCATCCCCACGGTGATGGGGGCGGTCGACCTGCCGTACTCCAAGGTCGACGGCATCGACCTGATCGTCGATGGCTACCACGGCGAGGTCTACACCAACCCCTCCGCCGAGCTGGTGCGCCAGTACAGCGACGTGGTCGCCGAGGAGCGCGAGCTGAGCAAGGGCCTGGCGGCCCTGCGCGAGCTGCCCTGCGAGACCCTCGACGGCCACCGCATGCCGCTCTGGGTCAACACCGGCCTGCTCGCCGATGTCGCCCGCGCCCAGGAGCGTGGCGCCGAGGGCGTGGGCCTGTACCGCACCGAAGTGCCGTTCATGATCAACGACCGCTTCCCCAGCGAGAAGGAACAGTTGGCGATCTACCGCGAGCAGCTCAGTGCCTTCCACCCGCTGCCGGTGACCATGCGCACCCTGGATATCGGCGGCGACAAGGCGCTGTCCTACTTCCCGATCAAGGAAGACAACCCGTTCCTCGGCTGGCGCGGCATCCGCGTCACCCTCGACCACCCGGAGATCTTCCTGGTCCAGACCCGCGCCATGCTCAAGGCCAGCGAAGGACTGGACAACCTGCGCATCCTGCTGCCGATGATCTCCGGCACCCACGAGCTGGAAGAGGCCCTGCACCTGATCCACCGCGCCTGGGGCGAGGTGCGCGACGAGGGCGTGGACATCGCCATGCCGCCCATCGGCATGATGGTCGAGATTCCCGCCGCCGTGTACCAGACCCGCGAGCTGGCCCGCCAGGTCGACTTCCTTTCGGTCGGTTCGAACGACCTGACCCAGTACCTGCTGGCGGTCGACCGCAACAATCCGCGGGTCGCCGACCTCTACGACTACCTGCATCCGGCCGTTCTGCATGCGTTGAAGAAGGTGGTCGACGATGCCCACCTGGAAGGCAAGCCGGTGAGCATCTGCGGCGAGATGGCCGGCGATCCCGCGGCTGCCGTGCTGCTGATGGCGATGGGCTTCGACAGCCTGTCGATGAACGCCACCAACCTGCCCAAGGTGAAGTGGCTGCTGCGCCAGATCACCCTGGACAAGGCCCGGGACCTGCTCGGCCAGTTGCTCACCTTCGACAACCCGCAGGTCATCCACAGCTCGCTGCACCTGGCGTTGCGCAACCTCGGCCTGGGTCGCGTGATCAACCCGGCGGCTACCGTCCAGCCCTGA |
| Protein Properties |
| Protein Residues: |
759 |
| Protein Molecular Weight: |
83.6 kDa |
| Protein Theoretical pI: |
5.56 |
| Hydropathicity (GRAVY score): |
-0.047 |
| Charge at pH 7 (predicted): |
-14.7 |
| Protein Sequence: |
>PA0337
MLNTLRKIVQEVNSAKDLKAALGIIVQRVKEAMGTQVCSVYLLDTETQRFVLMATEGLNKRSIGKVSMAPSEGLVGLVGTREEPLNLENAAAHPRYRYFAETGEERYASFLGAPIIHHRRVMGVLVVQQKERRQFDEGEEAFLVTMSAQLAGVIAHAEATGSIRGLGKLGKGIQEAKFVGVPGAPGVGVGKAVVVLPPADLEVVPDKQVDDIDAEIALFKQALEGVRADMRALSSKLASQLRKEERALFDVYLMMLDDASIGNEVKRIIRTGQWAQGALRQVVMEHVQRFELMDDAYLRERASDVKDIGRRLLAYLQEERKQNLTYPEQTIIVSEELSPAMLGEVPEGRLVGLVSVLGSGNSHVAILARAMGIPTVMGAVDLPYSKVDGIDLIVDGYHGEVYTNPSAELVRQYSDVVAEERELSKGLAALRELPCETLDGHRMPLWVNTGLLADVARAQERGAEGVGLYRTEVPFMINDRFPSEKEQLAIYREQLSAFHPLPVTMRTLDIGGDKALSYFPIKEDNPFLGWRGIRVTLDHPEIFLVQTRAMLKASEGLDNLRILLPMISGTHELEEALHLIHRAWGEVRDEGVDIAMPPIGMMVEIPAAVYQTRELARQVDFLSVGSNDLTQYLLAVDRNNPRVADLYDYLHPAVLHALKKVVDDAHLEGKPVSICGEMAGDPAAAVLLMAMGFDSLSMNATNLPKVKWLLRQITLDKARDLLGQLLTFDNPQVIHSSLHLALRNLGLGRVINPAATVQP |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0337 and its homologs
|




