| Identification |
| Name: |
Enolase |
| Synonyms: |
- 2-phospho-D-glycerate hydro-lyase
- 2-phosphoglycerate dehydratase
|
| Gene Name: |
eno |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in magnesium ion binding |
| Specific Function: |
Catalyzes the reversible conversion of 2- phosphoglycerate into phosphoenolpyruvate. It is essential for the degradation of carbohydrates via glycolysis. It is also a component of the RNA degradosome, a multi-enzyme complex involved in RNA processing and messenger RNA degradation. Its interaction with RNase E is important for the turnover of mRNA, in particular on transcripts encoding enzymes of energy-generating metabolic routes. Its presence in the degradosome is required for the response to excess phosphosugar. May play a regulatory role in the degradation of specific RNAs, such as ptsG mRNA, therefore linking cellular metabolic status with post-translational gene regulation |
| Cellular Location: |
Cytoplasm, cytoskeleton. Secreted. Cell surface. |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
| | |
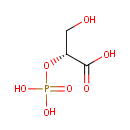
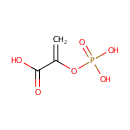 | → |  | + | 2-Phosphoglyceric acid | + | 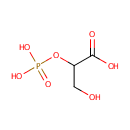 |
| | |
2-Phosphoglyceric acid | + |  | ↔ |  | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| macromolecular complex | | phosphopyruvate hydratase complex | | protein complex | | Function |
|---|
| binding | | carbon-oxygen lyase activity | | catalytic activity | | cation binding | | hydro-lyase activity | | ion binding | | lyase activity | | magnesium ion binding | | metal ion binding | | phosphopyruvate hydratase activity | | Process |
|---|
| alcohol metabolic process | | glucose catabolic process | | glucose metabolic process | | glycolysis | | hexose metabolic process | | metabolic process | | monosaccharide metabolic process | | small molecule metabolic process |
|
| Gene Properties |
| Locus tag: |
PA3635 |
| Strand: |
- |
| Entrez Gene ID: |
880453 |
| Accession: |
NP_252325.1 |
| GI: |
15598831 |
| Sequence start: |
4068677 |
| Sequence End: |
4069966 |
| Sequence Length: |
1289 |
| Gene Sequence: |
>PA3635
ATGGCAAAGATCGTCGACATCAAGGGCCGTGAGGTCCTGGACTCCCGCGGCAACCCTACCGTTGAAGCGGACGTGATCCTGGACAACGGCATCGTCGGCAGCGCCTGCGCGCCTTCCGGTGCTTCCACCGGTTCCCGCGAGGCCCTCGAGCTGCGCGATGGCGACAAGAGCCGTTACCTGGGCAAGGGCGTGCTGAAAGCCGTGGCCAACATCAACGGCCCGATCCGCGACCTGCTGCTGGGCAAGGACGCGGCCGACCAGAAAGCCCTCGACCACGCGATGATCGAGCTGGACGGCACCGAGAACAAGGCCAAGCTGGGCGCCAACGCGATCCTCGCGGTGTCCCTGGCTGCCGCCAAGGCCGCCGCACAGGCCAAGGGCGTACCGCTGTACGCGCACATCGCCGACCTCAACGGCACTCCCGGCCAGTACTCCATGCCGGTGCCGATGATGAACATCATCAACGGCGGCGAGCATGCGGACAACAACGTCGATATCCAGGAATTCATGGTCCAGCCGGTCGGCGCGAAGAACTTCGCCGAGGCGCTGCGCATGGGCGCCGAGATCTTCCATCACCTCAAGGCCGTGCTGAAGGCCCGTGGCCTGAACACCGCCGTCGGTGACGAAGGCGGCTTCGCGCCGAACCTGTCGTCCAACGAAGACGCCCTGGCCGCCATCGCCGAGGCCGTCGAGAAGGCCGGCTACAAGCTGGGCGACGACGTGACCCTGGCCCTGGACTGCGCCTCCAGCGAGTTCTTCAAGGACGGCAAGTACGACCTGGAAGGCGAAGGCAAGGTATTCGACGCCGCCGGTTTCGCCGACTACCTGGCCGGCCTGACCCAGCGCTACCCGATCATCTCCATCGAGGACGGCATGGACGAGTCCGACTGGGCCGGCTGGAAAGGCCTGACCGACAAGATCGGCGCCAAGGTCCAACTGGTCGGCGACGACCTGTTCGTGACCAACACCAAGATCCTCAAGGAAGGCATCGAGAAGGGCATCGGCAATTCGATCCTGATCAAGTTCAACCAGATCGGTTCGCTCACCGAGACCCTGGAGGCCATCCAGATGGCCAAGGCCGCCGGCTATACCGCGGTGATCTCGCACCGCTCCGGCGAAACCGAGGACTCGACCATCGCCGACCTGGCCGTGGGTACCGCCGCCGGTCAGATCAAGACCGGTTCGCTGTGCCGCTCCGACCGCGTGTCCAAGTACAACCAGTTGCTGCGCATCGAAGAGCAACTGGGCGCCAAGGCGCCGTACCGTGGTCGCGCGGAATTCCGCGGCTGA |
| Protein Properties |
| Protein Residues: |
429 |
| Protein Molecular Weight: |
45.2 kDa |
| Protein Theoretical pI: |
4.81 |
| Hydropathicity (GRAVY score): |
-0.142 |
| Charge at pH 7 (predicted): |
-12.63 |
| Protein Sequence: |
>PA3635
MAKIVDIKGREVLDSRGNPTVEADVILDNGIVGSACAPSGASTGSREALELRDGDKSRYLGKGVLKAVANINGPIRDLLLGKDAADQKALDHAMIELDGTENKAKLGANAILAVSLAAAKAAAQAKGVPLYAHIADLNGTPGQYSMPVPMMNIINGGEHADNNVDIQEFMVQPVGAKNFAEALRMGAEIFHHLKAVLKARGLNTAVGDEGGFAPNLSSNEDALAAIAEAVEKAGYKLGDDVTLALDCASSEFFKDGKYDLEGEGKVFDAAGFADYLAGLTQRYPIISIEDGMDESDWAGWKGLTDKIGAKVQLVGDDLFVTNTKILKEGIEKGIGNSILIKFNQIGSLTETLEAIQMAKAAGYTAVISHRSGETEDSTIADLAVGTAAGQIKTGSLCRSDRVSKYNQLLRIEEQLGAKAPYRGRAEFRG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3635 and its homologs
|