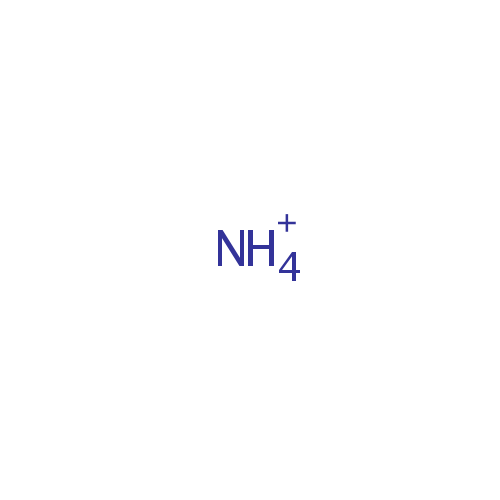
Ammonium (PAMDB001595)
Enzymes
- General function:
- Involved in oxidoreductase activity
- Specific function:
- L-glutamate + H(2)O + NADP(+) = 2-oxoglutarate + NH(3) + NADPH
- Gene Name:
- gdhA
- Locus Tag:
- PA4588
- Molecular weight:
- 48.9 kDa
Reactions
| L-glutamate + H(2)O + NADP(+) = 2-oxoglutarate + NH(3) + NADPH. |
- General function:
- Involved in asparaginase activity
- Specific function:
- L-asparagine + H(2)O = L-aspartate + NH(3)
- Gene Name:
- ansB
- Locus Tag:
- PA1337
- Molecular weight:
- 38.6 kDa
Reactions
| L-asparagine + H(2)O = L-aspartate + NH(3). |
- General function:
- Involved in cyanate hydratase activity
- Specific function:
- Catalyzes the reaction of cyanate with bicarbonate to produce ammonia and carbon dioxide
- Gene Name:
- cynS
- Locus Tag:
- PA2052
- Molecular weight:
- 16.8 kDa
Reactions
| Cyanate + HCO(3)(-) + 2 H(+) = NH(3) + 2 CO(2). |
- General function:
- Involved in D-serine ammonia-lyase activity
- Specific function:
- D-serine = pyruvate + NH(3)
- Gene Name:
- dsdA
- Locus Tag:
- PA3357
- Molecular weight:
- 48.2 kDa
Reactions
| D-serine = pyruvate + NH(3). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the formation of alpha-ketobutyrate from threonine in a two-step reaction. The first step is a dehydration of threonine, followed by rehydration and liberation of ammonia. Deaminates L-threonine, but also L-serine to a lesser extent
- Gene Name:
- ilvA
- Locus Tag:
- PA1326
- Molecular weight:
- 55.9 kDa
Reactions
| L-threonine = 2-oxobutanoate + NH(3). |
- General function:
- Involved in biosynthetic process
- Specific function:
- Catalyzes the biosynthesis of 4-amino-4-deoxychorismate (ADC) from chorismate and glutamine
- Gene Name:
- pabB
- Locus Tag:
- PA1758
- Molecular weight:
- 49.6 kDa
Reactions
| Chorismate + L-glutamine = 4-amino-4-deoxychorismate + L-glutamate. |
- General function:
- Involved in hydroxymethylbilane synthase activity
- Specific function:
- Tetrapolymerization of the monopyrrole PBG into the hydroxymethylbilane pre-uroporphyrinogen in several discrete steps
- Gene Name:
- hemC
- Locus Tag:
- PA5260
- Molecular weight:
- 33.6 kDa
Reactions
| 4 porphobilinogen + H(2)O = hydroxymethylbilane + 4 NH(3). |
- General function:
- Involved in oxidation-reduction process
- Specific function:
- Ammonium hydroxide + 3 NAD(P)(+) + H(2)O = nitrite + 3 NAD(P)H
- Gene Name:
- nirB
- Locus Tag:
- PA1781
- Molecular weight:
- 89.7 kDa
Reactions
| Ammonium hydroxide + 3 NAD(P)(+) + H(2)O = nitrite + 3 NAD(P)H. |
- General function:
- Involved in D-amino-acid dehydrogenase activity
- Specific function:
- Oxidative deamination of D-amino acids
- Gene Name:
- dadA
- Locus Tag:
- PA5304
- Molecular weight:
- 47.1 kDa
Reactions
| A D-amino acid + H(2)O + acceptor = a 2-oxo acid + NH(3) + reduced acceptor. |
- General function:
- Involved in glutaminase activity
- Specific function:
- L-glutamine + H(2)O = L-glutamate + NH(3)
- Gene Name:
- glsA2
- Locus Tag:
- PA1638
- Molecular weight:
- 33 kDa
Reactions
| L-glutamine + H(2)O = L-glutamate + NH(3). |
- General function:
- Involved in asparaginase activity
- Specific function:
- L-asparagine + H(2)O = L-aspartate + NH(3)
- Gene Name:
- ansA
- Locus Tag:
- PA2253
- Molecular weight:
- 34.8 kDa
Reactions
| L-asparagine + H(2)O = L-aspartate + NH(3). |
- General function:
- Involved in glutamate-ammonia ligase activity
- Specific function:
- ATP + L-glutamate + NH(3) = ADP + phosphate + L-glutamine
- Gene Name:
- glnA
- Locus Tag:
- PA5119
- Molecular weight:
- 51.9 kDa
Reactions
| ATP + L-glutamate + NH(3) = ADP + phosphate + L-glutamine. |
- General function:
- Involved in nitrite reductase [NAD(P)H] activity
- Specific function:
- Required for activity of the reductase
- Gene Name:
- nirD
- Locus Tag:
- PA1780
- Molecular weight:
- 11.6 kDa
Reactions
| Ammonium hydroxide + 3 NAD(P)(+) + H(2)O = nitrite + 3 NAD(P)H. |
- General function:
- Involved in aspartate ammonia-lyase activity
- Specific function:
- L-aspartate = fumarate + NH(3)
- Gene Name:
- aspA
- Locus Tag:
- PA5429
- Molecular weight:
- 51.1 kDa
Reactions
| L-aspartate = fumarate + NH(3). |
- General function:
- Involved in catalytic activity
- Specific function:
- Ethanolamine = acetaldehyde + NH(3)
- Gene Name:
- eutB
- Locus Tag:
- PA4024
- Molecular weight:
- 50.2 kDa
Reactions
| Ethanolamine = acetaldehyde + NH(3). |
- General function:
- Involved in L-serine ammonia-lyase activity
- Specific function:
- Deaminates also threonine, particularly when it is present in high concentration
- Gene Name:
- sdaA
- Locus Tag:
- PA2443
- Molecular weight:
- 48.9 kDa
Reactions
| L-serine = pyruvate + NH(3). |
- General function:
- Involved in NAD+ synthase (glutamine-hydrolyzing) activity
- Specific function:
- Catalyzes a key step in NAD biosynthesis, transforming deamido-NAD into NAD by a two-step reaction
- Gene Name:
- nadE
- Locus Tag:
- PA4920
- Molecular weight:
- 29.7 kDa
Reactions
| ATP + deamido-NAD(+) + NH(3) = AMP + diphosphate + NAD(+). |
- General function:
- Involved in hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
- Specific function:
- Cytosine + H(2)O = uracil + NH(3)
- Gene Name:
- codA
- Locus Tag:
- PA0437
- Molecular weight:
- 47.1 kDa
Reactions
| Cytosine + H(2)O = uracil + NH(3). |
- General function:
- Involved in zinc ion binding
- Specific function:
- Converts 2,5-diamino-6-(ribosylamino)-4(3h)-pyrimidinone 5'-phosphate into 5-amino-6-(ribosylamino)-2,4(1h,3h)- pyrimidinedione 5'-phosphate
- Gene Name:
- ribD
- Locus Tag:
- PA4056
- Molecular weight:
- 39.8 kDa
Reactions
| 2,5-diamino-6-hydroxy-4-(5-phosphoribosylamino)pyrimidine + H(2)O = 5-amino-6-(5-phosphoribosylamino)uracil + NH(3). |
| 5-amino-6-(5-phospho-D-ribitylamino)uracil + NADP(+) = 5-amino-6-(5-phospho-D-ribosylamino)uracil + NADPH. |
- General function:
- Involved in aminomethyltransferase activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine
- Gene Name:
- gcvT
- Locus Tag:
- PA5215
- Molecular weight:
- 38.9 kDa
Reactions
| [Protein]-S(8)-aminomethyldihydrolipoyllysine + tetrahydrofolate = [protein]-dihydrolipoyllysine + 5,10-methylenetetrahydrofolate + NH(3). |
- General function:
- Involved in L-serine ammonia-lyase activity
- Specific function:
- Deaminates also threonine, particularly when it is present in high concentration
- Gene Name:
- sdaB
- Locus Tag:
- PA5379
- Molecular weight:
- 49.2 kDa
Reactions
| L-serine = pyruvate + NH(3). |
- General function:
- Involved in glycine dehydrogenase (decarboxylating) activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The P protein binds the alpha-amino group of glycine through its pyridoxal phosphate cofactor; CO(2) is released and the remaining methylamine moiety is then transferred to the lipoamide cofactor of the H protein
- Gene Name:
- gcvP
- Locus Tag:
- PA5213
- Molecular weight:
- 104.7 kDa
Reactions
| Glycine + H-protein-lipoyllysine = H-protein-S-aminomethyldihydrolipoyllysine + CO(2). |
- General function:
- Involved in cellular amino acid biosynthetic process
- Specific function:
- ATP + NH(3) + CO(2) = ADP + carbamoyl phosphate
- Gene Name:
- arcC
- Locus Tag:
- PA5173
- Molecular weight:
- 33.1 kDa
Reactions
| ATP + NH(3) + CO(2) = ADP + carbamoyl phosphate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Involved in the breakdown of putrescine via the oxidation of L-glutamylputrescine
- Gene Name:
- puuB
- Locus Tag:
- PA2776
- Molecular weight:
- 46.7 kDa
Reactions
| Gamma-glutamylputrescine + H(2)O + O(2) = Gamma-glutamyl-gamma-aminobutyraldehyde + NH(3) + H(2)O(2). |
- General function:
- Involved in N-succinylarginine dihydrolase activity
- Specific function:
- Catalyzes the hydrolysis of N(2)-succinylarginine into N(2)-succinylornithine, ammonia and CO(2)
- Gene Name:
- astB
- Locus Tag:
- PA0899
- Molecular weight:
- 48.9 kDa
Reactions
| N(2)-succinyl-L-arginine + 2 H(2)O = N(2)-succinyl-L-ornithine + 2 NH(3) + CO(2). |
- General function:
- Involved in hydrolase activity
- Specific function:
- Catalyzes the hydrolytic deamination of guanine, producing xanthine and ammonia
- Gene Name:
- guaD
- Locus Tag:
- PA1521
- Molecular weight:
- 48.3 kDa
Reactions
| Guanine + H(2)O = xanthine + NH(3). |
- General function:
- Involved in ureidoglycolate hydrolase activity
- Specific function:
- Involved in the anaerobic utilization of allantoin. Reinforces the induction of genes involved in the degradation of allantoin and glyoxylate by producing glyoxylate
- Gene Name:
- allA
- Locus Tag:
- PA1514
- Molecular weight:
- 19 kDa
Reactions
| (S)-ureidoglycolate + H(2)O = glyoxylate + 2 NH(3) + CO(2). |
- General function:
- Amino acid transport and metabolism
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The H protein shuttles the methylamine group of glycine from the P protein to the T protein
- Gene Name:
- gcvH
- Locus Tag:
- PA5214
- Molecular weight:
- 13.6 kDa
- General function:
- Coenzyme transport and metabolism
- Specific function:
- Catalyzes the oxidation of either pyridoxine 5'- phosphate (PNP) or pyridoxamine 5'-phosphate (PMP) into pyridoxal 5'-phosphate (PLP)
- Gene Name:
- pdxH
- Locus Tag:
- PA1049
- Molecular weight:
- 24.9 kDa
Reactions
| Pyridoxamine 5'-phosphate + H(2)O + O(2) = pyridoxal 5'-phosphate + NH(3) + H(2)O(2). |
| Pyridoxine 5'-phosphate + O(2) = pyridoxal 5'-phosphate + H(2)O(2). |
- General function:
- pyridine nucleotide biosynthetic process
- Specific function:
- Has nicotinamidemononucleotide (NMN) aminohydrolase activity, not active on other substrates.
- Gene Name:
- pncC
- Locus Tag:
- PA3618
- Molecular weight:
- 17.7 kDa
Reactions
| Beta-nicotinamide D-ribonucleotide + H(2)O = beta-nicotinate D-ribonucleotide + NH(3) |
Transporters
- General function:
- Involved in ammonium transmembrane transporter activity
- Specific function:
- Involved in the uptake of ammonia
- Gene Name:
- amtB
- Locus Tag:
- PA5287
- Molecular weight:
- 45.8 kDa