| Identification |
| Name: |
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase |
| Synonyms: |
- Meso-A2pm-adding enzyme
- Meso-diaminopimelate-adding enzyme
- UDP-MurNAc-L-Ala-D-Glu:meso-diaminopimelate ligase
- UDP-MurNAc-tripeptide synthetase
- UDP-N-acetylmuramyl-tripeptide synthetase
|
| Gene Name: |
murE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in ATP binding |
| Specific Function: |
Catalyzes the addition of meso-diaminopimelic acid to the nucleotide precursor UDP-N-acetylmuramoyl-L-alanyl-D-glutamate (UMAG) in the biosynthesis of bacterial cell-wall peptidoglycan. Is also able to use many meso-diaminopimelate analogs as substrates, although much less efficiently, but not L-lysine |
| Cellular Location: |
Cytoplasm (Probable) |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
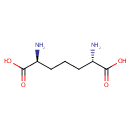
UDP-N-acetylmuramoyl-L-alanyl-D-glutamate | + | 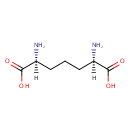 | + |  | + |  | → | Adenosine diphosphate | + | 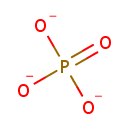 | + |  | + | 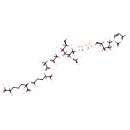 | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
meso-diaminopimelate | + |  | + |  | → |  | + | 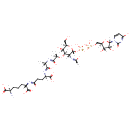 | + |  | + |  |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB000148 | Adenosine triphosphate | MetaboCard | | PAMDB000345 | ADP | MetaboCard | | PAMDB000360 | Diaminopimelic acid | MetaboCard | | PAMDB001633 | Hydrogen ion | MetaboCard | | PAMDB001776 | Inorganic phosphate | MetaboCard | | PAMDB002055 | Meso-2,6-Diaminoheptanedioate | MetaboCard | | PAMDB000383 | Phosphate | MetaboCard | | PAMDB001049 | UDP-N-Acetylmuramoyl-L-alanyl-D-gamma-glutamyl-meso-2,6-diaminopimelate | MetaboCard | | PAMDB000649 | UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate | MetaboCard | | PAMDB000650 | UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate | MetaboCard |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| acid-amino acid ligase activity | | adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | catalytic activity | | ligase activity | | ligase activity, forming carbon-nitrogen bonds | | nucleoside binding | | purine nucleoside binding | | Process |
|---|
| aminoglycan metabolic process | | biological regulation | | biosynthetic process | | carbohydrate metabolic process | | cell division | | cell wall biogenesis | | cell wall organization or biogenesis | | cellular cell wall organization or biogenesis | | cellular component organization or biogenesis | | cellular process | | glycosaminoglycan metabolic process | | metabolic process | | peptidoglycan biosynthetic process | | peptidoglycan metabolic process | | peptidoglycan-based cell wall biogenesis | | polysaccharide metabolic process | | primary metabolic process | | regulation of biological quality | | regulation of cell shape |
|
| Gene Properties |
| Locus tag: |
PA4417 |
| Strand: |
- |
| Entrez Gene ID: |
881246 |
| Accession: |
NP_253107.1 |
| GI: |
15599613 |
| Sequence start: |
4951141 |
| Sequence End: |
4952604 |
| Sequence Length: |
1463 |
| Gene Sequence: |
>PA4417
ATGCCTATGAGCCTGAACCAACTGTTTCCCCAGGCCGAGCGCGATCTGCTGATCCGCGAGCTGACCCTGGATAGCCGCGGCGTTCGTCCGGGCGACCTGTTCCTGGCGGTGCCGGGCGGGCGCCAGGATGGTCGTGCGCACATCGCCGATGCCCTGGCCAAGGGCGCGGCTGCCGTGGCTTACGAGGCGGAAGGCGCCGGAGAGTTGCCGCCCAGCGATGCGCCGCTGATCGCGGTGAAGGGGCTGGCCGCGCAACTGTCGGCGGTCGCCGGGCGTTTCTACGGCGAGCCGAGCCGCGGGCTGGACCTGATCGGCGTCACCGGCACCAACGGCAAGACCAGCGTCAGCCAACTGGTGGCCCAGGCCCTGGATCTGCTCGGCGAGCGCTGCGGCATCGTCGGCACCCTCGGCACCGGTTTCTACGGCGCCCTGGAGAGCGGCCGGCACACCACGCCGGACCCGCTCGCGGTGCAGGCCACGCTGGCCACGCTGAAGCAGGCCGGCGCCCGCGCGGTAGCGATGGAAGTGTCTTCCCACGGCCTCGACCAGGGCCGCGTGGCGGCGCTCGGTTTCGATATCGCGGTGTTCACCAATCTGTCCCGCGACCACCTCGACTATCACGGTTCGATGGAAGCCTATGCCGCCGCCAAGGCCAAGCTGTTCGCCTGGCCGGGCCTGCGCTGCCGGGTGATCAACCTGGACGACGATTTCGGCCGTCGACTGGCCGGCGAGGAGCAGGACTCGGAGCTGATCACCTACAGCCTCACCGACAGCTCGGCGTTCCTCTATTGCCGCGAAGCGCGCTTCGGCGACGCCGGCATCGAGGCGGCGCTGGTCACTCCGCACGGCGAGGGCCTGCTGCGCAGCCCGTTGCTCGGCCGCTTCAACCTGAGCAACCTGCTGGCGGCGGTCGGTGCGTTGCTTGGCCTGGGTTATCCCCTGGGCGATATCCTCCGCACTTTGCCGCAACTGCAGGGGCCGGTCGGCCGCATGCAGCGCCTGGGAGGCGGCGACAAGCCGCTGGTGGTGGTGGACTACGCGCATACTCCCGACGCCCTGGAAAAAGTCCTGGAGGCCCTGCGTCCGCACGCGGCCGCGCGCCTGCTGTGCCTGTTCGGCTGCGGTGGCGATCGCGATGCCGGCAAGCGTCCGCTGATGGCTGCGATCGCCGAACGCCTGGCGGATGAGGTGCTGGTCACCGACGACAACCCGCGCACCGAGGCCAGTGCGGCGATCATCGCCGATATCCGCAAAGGCTTCGCTGCCGCTGACAAGGTTACCTTCCTGCCGTCGCGCGGTGAGGCGATCGCCCATCTGATCGCTTCCGCTGCGGTGGATGACGTGGTGCTCCTGGCCGGCAAGGGTCACGAGGATTATCAGGAGATCGACGGCGTACGCCATCCGTTCTCCGACATCGAGCAGGCCGAGCGCGCCCTGGCCGCCTGGGAGGTGCCGCATGCTTGA |
| Protein Properties |
| Protein Residues: |
487 |
| Protein Molecular Weight: |
51.3 kDa |
| Protein Theoretical pI: |
5.11 |
| Hydropathicity (GRAVY score): |
0.044 |
| Charge at pH 7 (predicted): |
-13.24 |
| Protein Sequence: |
>PA4417
MPMSLNQLFPQAERDLLIRELTLDSRGVRPGDLFLAVPGGRQDGRAHIADALAKGAAAVAYEAEGAGELPPSDAPLIAVKGLAAQLSAVAGRFYGEPSRGLDLIGVTGTNGKTSVSQLVAQALDLLGERCGIVGTLGTGFYGALESGRHTTPDPLAVQATLATLKQAGARAVAMEVSSHGLDQGRVAALGFDIAVFTNLSRDHLDYHGSMEAYAAAKAKLFAWPGLRCRVINLDDDFGRRLAGEEQDSELITYSLTDSSAFLYCREARFGDAGIEAALVTPHGEGLLRSPLLGRFNLSNLLAAVGALLGLGYPLGDILRTLPQLQGPVGRMQRLGGGDKPLVVVDYAHTPDALEKVLEALRPHAAARLLCLFGCGGDRDAGKRPLMAAIAERLADEVLVTDDNPRTEASAAIIADIRKGFAAADKVTFLPSRGEAIAHLIASAAVDDVVLLAGKGHEDYQEIDGVRHPFSDIEQAERALAAWEVPHA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4417 and its homologs
|