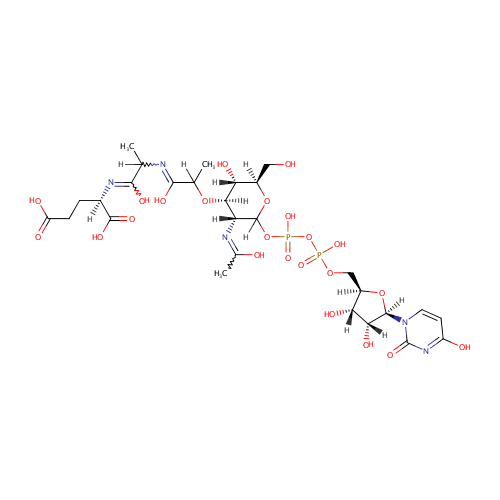
UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate (PAMDB000649)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000649 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | UDP-n-acetylmuramoyl-L-alanyl-D-glutamate is a member of the chemical class known as Peptides. These are compounds containing an amide derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another. UDP-N-acetylmuramoyl-L-alanyl-D-glutamate is a key component of peptidoglycan synthesis. The peptidoglycan synthesis pathway starts at the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-alpha-D-glucosamine, yielding the complete monomeric unit a lipid , also known as lipid . This final lipid intermediate is transferred through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. UDP-N-acetylmuramoyl-L-alanyl-D-glutamate:meso-diaminopimelate ligase is a cytoplasmic enzyme that catalyzes the addition of meso-diaminopimelic acid to nucleotide precursor UDP-N-acetylmuramoyl-L-alanyl-D-glutamate in the biosynthesis of bacterial cell-wall peptidoglycan. (PMID 11124264) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C22H32Cl2N2O4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 459.41 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 458.1739129 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | IEKOTSCYBBDIJC-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C22H32Cl2N2O4/c1-3-5-7-13-26(14-8-6-4-2)22(30)19(11-12-20(27)28)25-21(29)16-9-10-17(23)18(24)15-16/h9-10,15,19H,3-8,11-14H2,1-2H3,(H,25,29)(H,27,28) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 17088-64-1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2R)-2-({2-[(2-{[(3R,4R,5S,6R)-2-({[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-5-hydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-4-yl]oxy}-1-hydroxypropylidene)amino]-1-hydroxypropylidene}amino)pentanedioic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (2R)-2-({2-[(2-{[(3R,4R,5S,6R)-2-[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]-5-hydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-4-yl]oxy}-1-hydroxypropylidene)amino]-1-hydroxypropylidene}amino)pentanedioic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CCCCCN(CCCCC)C(=O)C(CCC(O)=O)N=C(O)C1=CC(Cl)=C(Cl)C=C1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrimidine nucleotides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrimidine nucleotide sugars | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrimidine nucleotide sugars | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Diaminopimelic acid + Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate > ADP + Hydrogen ion + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate Adenosine triphosphate + D-Glutamic acid + UDP-N-Acetylmuramoyl-L-alanine <> ADP + Hydrogen ion + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanine + D-Glutamic acid <> ADP + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate + Meso-2,6-Diaminoheptanedioate <> ADP + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate <i>meso</i>-diaminopimelate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate + Adenosine triphosphate > Hydrogen ion + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate + Phosphate + ADP UDP-N-Acetylmuramoyl-L-alanine + Adenosine triphosphate + D-Glutamic acid > Adenosine diphosphate + Phosphate + UDP-N-acetylmuramoyl-L-alanyl-D-glutamate + ADP + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate UDP-N-acetylmuramoyl-L-alanyl-D-glutamate + Meso-2,6-Diaminoheptanedioate + Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamate > Adenosine diphosphate + Phosphate + Hydrogen ion + UDP-N-Acetylmuramoyl-L-alanyl-D-gamma-glutamyl-meso-2,6-diaminopimelate + ADP | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||