| Identification |
| Name: |
Lipopolysaccharide export system protein lptA |
| Synonyms: |
Not Available |
| Gene Name: |
lptA |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in lipopolysaccharide binding |
| Specific Function: |
Required for the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane. May act as a chaperone that facilitates LPS transfer across the aquaeous environment of the periplasm. Interacts specifically with the lipid A domain of LPS |
| Cellular Location: |
Periplasm |
| KEGG Pathways: |
- Lipopolysaccharide biosynthesis PW000831
- superpathway of phospholipid biosynthesis I (bacteria) PHOSLIPSYN-PWY
- CDP-diacylglycerol biosynthesis II PWY0-1319
- phosphatidylglycerol biosynthesis I (plastidic) PWY4FS-7
- phosphatidylglycerol biosynthesis II (non-plastidic) PWY4FS-8
- superpathway phosphatidate biosynthesis (yeast) PWY-7411
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
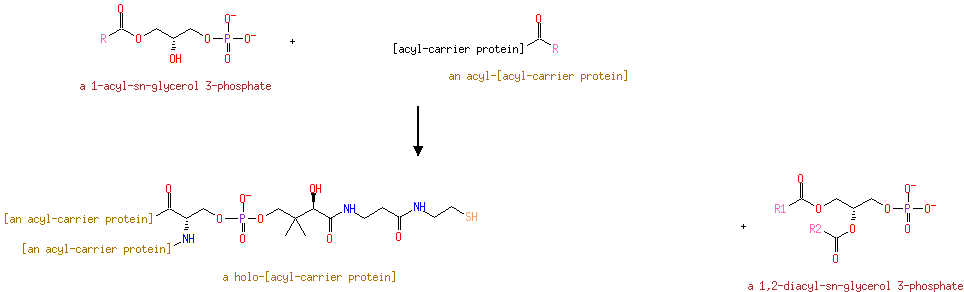
| a 1-acyl- sn-glycerol 3-phosphate + an acyl-[acyl-carrier protein] → a holo-[acyl-carrier protein] + a 1,2-diacyl- sn-glycerol 3-phosphate ReactionCard | |
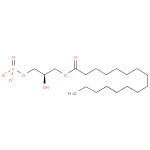 | → | 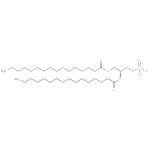 |
| | |
Saturated-2-Lysophosphatidates | → | 2-3-4-Saturated-L-Phosphatidates |
| Saturated-2-Lysophosphatidates → 2-3-4-Saturated-L-Phosphatidates ReactionCard | | |
|
| Complex Reactions: |
Not Available |
| Transports: |
|
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | periplasmic space | | Function |
|---|
| binding | | lipopolysaccharide binding | | Process |
|---|
| carbohydrate transport | | establishment of localization | | lipopolysaccharide transport | | polysaccharide transport | | transport |
|
| Gene Properties |
| Locus tag: |
PA0005 |
| Strand: |
- |
| Entrez Gene ID: |
877576 |
| Accession: |
NP_064725.1 |
| GI: |
15595203 |
| Sequence start: |
7018 |
| Sequence End: |
7791 |
| Sequence Length: |
773 |
| Gene Sequence: |
>PA0005
ATGTCGACAGTGCAGGCCATCAGAACCGTCCTCTTTTACCTGCTGCTGTCCGCCAGCGCGTTCGTCTGGGGCACCCTCAGCTTCTTCATCGCGCCGATCCTGCCGTTCCGCGCCCGCTACCGCTTCGTGGTACAGAACTGGTGCCGCTTCGCGATCTGGCTGACCCGCGTGGTCGCCGGCATCCGCTACGAGGTGCGCGGACTGGAGAACATCCCGGAAAAGCCCTGCGTGATCCTCTCCAAGCACCAGAGCACCTGGGAAACCTTCTTCCTCTCCGGCTTCTTCGAGCCACTCAGCCAGGTACTCAAGCGCGAGCTGCTCTACGTGCCGTTCTTCGGCTGGGCCCTGGCCCTGCTCAAGCCCATCGCCATCGACCGCAGCCAGCCCAAGCTGGCCCTCAAGCAACTGGCCAAGCAGGGCGACGAGTGCCTGAAGAAAGGCGCCTGGGTGCTGATCTTCCCGGAAGGCACGCGTATTCCGGTGGGGCAGATGGGCAAGTTCTCCCGCGGCGGCACCGCCCTGGCGGTCAACGCCGGGCTACCGGTACTGCCGATCGCCCACAACGCCGGGCAGTATTGGCCCAAGGCCGGCTGGGCCAAGTACCCGGGCACCATCCAGGTGGTGATCGGCCCGGCCATGCACGCCGAAGGCGAAGGCCCGCGCGCCATCGCCGAGCTAAACCAGCGCGCCGAAGCCTGGGTCAGCGAGACCATGGCCGAGATCAGCCCCATCCAGCAGCGGGTCAGCCATCCGGAGCCGTCGGTGGTCTCGTGA |
| Protein Properties |
| Protein Residues: |
257 |
| Protein Molecular Weight: |
28.7 kDa |
| Protein Theoretical pI: |
10.23 |
| Hydropathicity (GRAVY score): |
0.182 |
| Charge at pH 7 (predicted): |
10.86 |
| Protein Sequence: |
>PA0005
MSTVQAIRTVLFYLLLSASAFVWGTLSFFIAPILPFRARYRFVVQNWCRFAIWLTRVVAGIRYEVRGLENIPEKPCVILSKHQSTWETFFLSGFFEPLSQVLKRELLYVPFFGWALALLKPIAIDRSQPKLALKQLAKQGDECLKKGAWVLIFPEGTRIPVGQMGKFSRGGTALAVNAGLPVLPIAHNAGQYWPKAGWAKYPGTIQVVIGPAMHAEGEGPRAIAELNQRAEAWVSETMAEISPIQQRVSHPEPSVVS |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0005 and its homologs
|