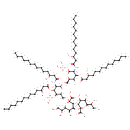
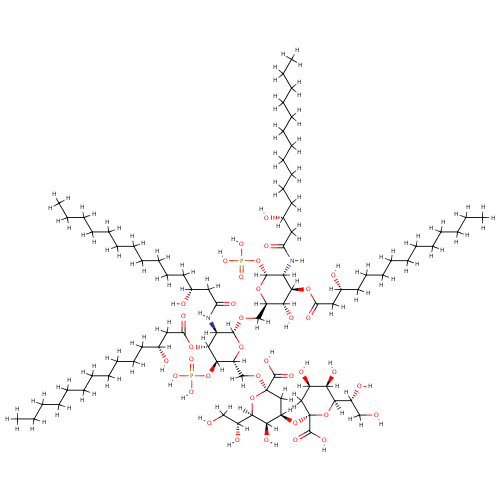
KDO(2)-lipid IV(A) (PAMDB001643)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB001643 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | KDO(2)-lipid IV(A) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Kdo(2)-lipid iv(a) belongs to the class of Polysaccharide Phosphates. These are polysaccharides in which a phosphate group is bound to at least one carbohydrate unit. (inferred from compound structure) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C84H154N2O37P2 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 1846.0603 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 1844.970566976 | ||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | XAOLJGCZESYRFT-VHSKNIDJSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C84H154N2O37P2/c1-5-9-13-17-21-25-29-33-37-41-55(89)45-65(96)85-69-77(117-67(98)47-57(91)43-39-35-31-27-23-19-15-11-7-3)73(102)63(115-80(69)123-125(110,111)112)53-113-79-70(86-66(97)46-56(90)42-38-34-30-26-22-18-14-10-6-2)78(118-68(99)48-58(92)44-40-36-32-28-24-20-16-12-8-4)76(122-124(107,108)109)64(116-79)54-114-83(81(103)104)50-62(72(101)75(120-83)61(95)52-88)119-84(82(105)106)49-59(93)71(100)74(121-84)60(94)51-87/h55-64,69-80,87-95,100-102H,5-54H2,1-4H3,(H,85,96)(H,86,97)(H,103,104)(H,105,106)(H2,107,108,109)(H2,110,111,112)/t55-,56-,57-,58-,59-,60-,61-,62-,63-,64-,69-,70-,71-,72-,73-,74-,75-,76-,77-,78-,79-,80-,83-,84-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 143600-83-3 | ||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2R,4R,5R,6R)-4-{[(2R,4R,5R,6R)-2-carboxy-6-[(1R)-1,2-dihydroxyethyl]-4,5-dihydroxyoxan-2-yl]oxy}-6-[(1R)-1,2-dihydroxyethyl]-5-hydroxy-2-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-3-hydroxy-5-[(3R)-3-hydroxytetradecanamido]-4-{[(3R)-3-hydroxytetradecanoyl]oxy}-6-(phosphonooxy)oxan-2-yl]methoxy}-5-[(3R)-3-hydroxytetradecanamido]-4-{[(3R)-3-hydroxytetradecanoyl]oxy}-3-(phosphonooxy)oxan-2-yl]methoxy}oxane-2-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (kdo)2-lipid iva | ||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H]OC(=O)[C@]1(OC([H])([H])[C@@]2([H])O[C@@]([H])(OC([H])([H])[C@@]3([H])O[C@]([H])(OP(=O)(O[H])O[H])[C@]([H])(N([H])C(=O)C([H])([H])[C@]([H])(O[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H])[C@@]([H])(OC(=O)C([H])([H])[C@]([H])(O[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H])[C@]3([H])O[H])[C@]([H])(N([H])C(=O)C([H])([H])[C@]([H])(O[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H])[C@@]([H])(OC(=O)C([H])([H])[C@]([H])(O[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H])[C@]2([H])OP(=O)(O[H])O[H])O[C@]([H])([C@]([H])(O[H])C([H])([H])O[H])[C@]([H])(O[H])[C@]([H])(O[C@]2(O[C@]([H])([C@]([H])(O[H])C([H])([H])O[H])[C@]([H])(O[H])[C@]([H])(O[H])C2([H])[H])C(=O)O[H])C1([H])[H] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as saccharolipids. These are compounds in which fatty acids are linked directly to a sugar backbone, forming structures that are compatible with membrane bilayers. In the saccharolipids, a sugar substitutes for the glycerol backbone that is present in glycerolipids and glycerophospholipids. The most familiar saccharolipids contain an acylated glucosamine. In contrast to others glycolipids, the fatty acid is not glycosidically linked to the sugar moiety. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Saccharolipids | ||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Saccharolipids | ||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -6 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Membrane | ||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + KDO(2)-lipid IV(A) > ADP + Hydrogen ion + Phosphate + KDO(2)-lipid IV(A) Adenosine triphosphate + Water + KDO(2)-lipid IV(A) > ADP + Hydrogen ion + Phosphate + KDO(2)-lipid IV(A) -->-->Dodecanoyl-ACP (n-C12:0ACP) + KDO(2)-lipid IV(A) > acyl carrier protein + KDO(2)-lipid IV(A) with laurate cis-hexadec-9-enoyl-[acyl-carrier protein] (n-C16:1) + KDO(2)-lipid IV(A) > acyl carrier protein + KDO(2)-lipid IV(A) with palmitoleoyl CMP-3-Deoxy-D-manno-octulosonate + KDO-lipid IV(A) > Cytidine monophosphate + Hydrogen ion + KDO(2)-lipid IV(A) Lauroyl-KDO2-lipid IV(A) + Acyl-carrier protein + Dodecanoyl-[acyl-carrier protein] + KDO(2)-lipid IV(A) <> Dodecanoyl-[acyl-carrier protein] + Di[3-deoxy-D-manno-octulosonyl]-lipid IV(A) + Lauroyl-KDO2-lipid IV(A) + Acyl-carrier protein Hexadecenoyl-[acyl-carrier protein] + KDO(2)-lipid IV(A) <> (9Z)-Hexadec-9-enoyl-KDO2-lipid IV(A) + Acyl-carrier protein | ||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in biosynthetic process
- Specific function:
- Essential step in lipopolysaccharides biosynthesis. Acts at transfer of 3-deoxy-D-mono octulonic acid (KDO) from CMP-KDO to a tetraacyldisaccharide 1,4'-bisphosphate precursor of lipid A (lipid IVA). Transfers two molecules of KDO to lipid IVA. Degraded by FtsH; therefore FtsH regulates the addition of the sugar moiety of the LPS and thus the maturation of the LPS precursor
- Gene Name:
- waaA
- Locus Tag:
- PA4988
- Molecular weight:
- 46.5 kDa
- General function:
- Involved in transferase activity, transferring pentosyl groups
- Specific function:
- Catalyzes the transfer of the L-Ara4N moiety of the glycolipid undecaprenyl phosphate-alpha-L-Ara4N to lipid A. The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides
- Gene Name:
- arnT
- Locus Tag:
- PA3556
- Molecular weight:
- 61.7 kDa
Reactions
| 4-amino-4-deoxy-alpha-L-arabinopyranosyl di-trans,octa-cis-undecaprenyl phosphate + lipid IV(A) = lipid II(A) + di-trans,octa-cis-undecaprenyl phosphate. |
- General function:
- Involved in lipopolysaccharide transport
- Specific function:
- Part of the ABC transporter complex lptBFG involved in the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane
- Gene Name:
- lptG
- Locus Tag:
- PA3827
- Molecular weight:
- 39.2 kDa
- General function:
- Involved in lipopolysaccharide-transporting ATPase acti
- Specific function:
- Part of the ABC transporter complex lptBFG involved in the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane
- Gene Name:
- lptF
- Locus Tag:
- PA3828
- Molecular weight:
- 41.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Involved in lipid A export and possibly also in glycerophospholipid export and for biogenesis of the outer membrane. Transmembrane domains (TMD) form a pore in the inner membrane and the ATP-binding domain (NBD) is responsible for energy generation
- Gene Name:
- msbA
- Locus Tag:
- PA4997
- Molecular weight:
- 66.4 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex lptBFG involved in the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane. Probably responsible for energy coupling to the transport system
- Gene Name:
- lptB
- Locus Tag:
- PA4461
- Molecular weight:
- 26.5 kDa
- General function:
- Involved in lipopolysaccharide transmembrane transporter activity
- Specific function:
- Required for the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane
- Gene Name:
- lptC
- Locus Tag:
- PA4459
- Molecular weight:
- 21.4 kDa
- General function:
- Involved in lipopolysaccharide binding
- Specific function:
- Required for the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane. May act as a chaperone that facilitates LPS transfer across the aquaeous environment of the periplasm. Interacts specifically with the lipid A domain of LPS
- Gene Name:
- lptA
- Locus Tag:
- PA0005
- Molecular weight:
- 28.7 kDa
Transporters
- General function:
- Involved in lipopolysaccharide transport
- Specific function:
- Part of the ABC transporter complex lptBFG involved in the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane
- Gene Name:
- lptG
- Locus Tag:
- PA3827
- Molecular weight:
- 39.2 kDa
- General function:
- Involved in lipopolysaccharide-transporting ATPase acti
- Specific function:
- Part of the ABC transporter complex lptBFG involved in the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane
- Gene Name:
- lptF
- Locus Tag:
- PA3828
- Molecular weight:
- 41.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Involved in lipid A export and possibly also in glycerophospholipid export and for biogenesis of the outer membrane. Transmembrane domains (TMD) form a pore in the inner membrane and the ATP-binding domain (NBD) is responsible for energy generation
- Gene Name:
- msbA
- Locus Tag:
- PA4997
- Molecular weight:
- 66.4 kDa