| Identification |
| Name: |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase |
| Synonyms: |
- 4-amino-4-deoxy-L-arabinose lipid A transferase
- Polymyxin resistance protein pmrK
- Undecaprenyl phosphate-alpha-L-Ara4N transferase
|
| Gene Name: |
arnT |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in transferase activity, transferring pentosyl groups |
| Specific Function: |
Catalyzes the transfer of the L-Ara4N moiety of the glycolipid undecaprenyl phosphate-alpha-L-Ara4N to lipid A. The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides |
| Cellular Location: |
Cell inner membrane; Multi-pass membrane protein |
| KEGG Pathways: |
- Cationic antimicrobial peptide (CAMP) resistance pae01503
|
| KEGG Reactions: |
|
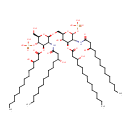
Undecaprenyl phosphate alpha-L-Ara4N | + | 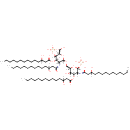 | ↔ | Lipid IIA | + |  |
| | |
Undecaprenyl phosphate alpha-L-Ara4N | + | Di[3-deoxy-D-manno-octulosonyl]-lipid IV(A) | ↔ | alpha-Kdo-(2->4)-alpha-Kdo-(2->6)-[4-P-L-Ara4N]-lipid IVA | + |  |
| | | |
|
| SMPDB Reactions: |
|
2 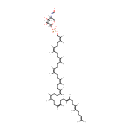 | + |  | → | 2 di-trans,octa-cis-undecaprenyl diphosphate | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
 | + | 4-amino-4-deoxy-α-L-arabinopyranosyl ditrans,octacis-undecaprenyl phosphate | → |  | + |  |
| |
|
| Complex Reactions: |
|
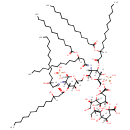
core oligosaccharide lipid A | + | 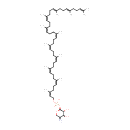 | → |  | + | 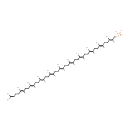 |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB006296 | (KDO)2-lipid A | MetaboCard | | PAMDB000906 | 2,3,2'3'-Tetrakis(3-hydroxytetradecanoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate | MetaboCard | | PAMDB001545 | 2,3,2'3'-Tetrakis(beta-hydroxymyristoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate | MetaboCard | | PAMDB003490 | 4-Amino-4-deoxy-alpha-L-arabinopyranosyl di-trans,octa-cis-undecaprenyl phosphate | MetaboCard | | PAMDB001892 | 4-Amino-4-deoxy-L-arabinose | MetaboCard | | PAMDB001582 | 4-Amino-4-deoxy-L-arabinose modified core oligosaccharide lipid A | MetaboCard | | PAMDB001145 | Di-trans,poly-cis-undecaprenyl phosphate | MetaboCard | | PAMDB001643 | KDO(2)-lipid IV(A) | MetaboCard | | PAMDB001447 | KDO2-Lipid A | MetaboCard | | PAMDB001379 | L-Ara4N-modified KDO2-Lipid A | MetaboCard | | PAMDB003413 | lipid II(A) | MetaboCard | | PAMDB005112 | Lipid IIA | MetaboCard | | PAMDB001054 | Undecaprenyl phosphate | MetaboCard | | PAMDB001760 | undecaprenyl phosphate-4-amino-4-deoxy-L-arabinose | MetaboCard |
|
| GO Classification: |
| Component |
|---|
| cell part | | membrane | | Function |
|---|
| catalytic activity | | mannosyltransferase activity | | transferase activity | | transferase activity, transferring glycosyl groups | | transferase activity, transferring hexosyl groups | | Process |
|---|
| macromolecule metabolic process | | macromolecule modification | | metabolic process | | protein amino acid glycosylation | | protein amino acid O-linked glycosylation | | protein modification process |
|
| Gene Properties |
| Locus tag: |
PA3556 |
| Strand: |
+ |
| Entrez Gene ID: |
879122 |
| Accession: |
NP_252246.1 |
| GI: |
15598752 |
| Sequence start: |
3984890 |
| Sequence End: |
3986539 |
| Sequence Length: |
1649 |
| Gene Sequence: |
>PA3556
ATGAGCCGCCGCCAGACCTGTTCGCTGCTGCTGATCGCCTTCGGCCTGTTCTACCTGGTTCCATTGAGCAACCATGGCCTGTGGATTCCCGACGAGACCCGCTACGCGCAGATCAGCCAGGCGATGCTGCTGGGCGGCGACTGGGTGTCGCCGCACTTCCTCGGCCTGCGCTACTTCGAGAAGCCGGTCGCCGGCTACTGGATGATCGCCCTCGGCCAGGCGGTCTTCGGCGAAAACCTGTTCGGCGTGCGCATCGCCTCGGTCGTCGCCACCGCCCTCAGCGTCCTGCTCGCCTACCTGCTGGCCCGCCGCCTGTGGCGCGACCCGCGCACCAGCCTGGCATGCGCGCTGCTCTACGCCAGCTTCGGCTTGATCGCCGGGCAGTCCGGCTATGCCAACCTCGACCCGCAATTCACCTTCTGGGTCAACCTGAGCCTGGTGGCGCTGTGGCATGCCCTCGACGCGGGGAGCCGCCGCGCGCGCCTGCTCGGCTGGACCCTGCTCGGCCTCGCCTGCGGCATGGGCTTCCTGACCAAGGGCTTCCTCGCCTGGCTGCTGCCGGTACTGGTCGCCCTGCCCTACATGCTCTGGCAGCGGCGCTGGCGCGAGCTGCTTGGCTACGGCGCGCTGGCGGTGCTGGCGGCGCTGCTCGTCTGCCTGCCCTGGGCGCTCGCCGTGCATGCACGGGAAGCGGACTACTGGCGGTTCTTCTTCTGGCACGAACACATCCGCCGCTTCGCCGGCGAAGACGCCCAGCACTCTCGCCCGTGGTGGTTCTACCTGCCGTTGCTGGCGGTCGCCTGCCTGCCCTGGAGCGGCCTGCTGCCGAGCGCTCTGCGCCAGGCCTGGCACGAGCGGCGCCAGGCGCCGGTGGTCTTCCTGGCGCTATGGCTGCTGTTGCCGCTGGCGTTCTTCAGCCTGAGCAGGGGCAAGCTGCCGACCTACATCATGCCCTGCCTGCTGCCGCTGGCACTGCTCATGGGCCACGCCCTGGTGCAGCGGCTGCGCCTGGGGAACAGCGTCGCGCTGCGCGGCAACGGGCTGCTCAACCTGGGCCTGGCGCTCCTCGCGCTGGCGGCCCTGGCCTACCTGCAACTGCGCAAGCCGGTGTACCAGGAAGAACCCTTCGAGCTGTTCCTGGTCCTGCTGGTGATCGGCGCCTGGGCCGCCGCCGGCCTCGCCCAGTGGCGCTACCCGCTACGCGCCTGGGCCGCGCCGCTGCTGGCGAGCTGGGTGCTGATCGCGCTGCTGCCGGCAGCGATGCCCAACCATGTGGTGCAGAACAAGACCCCCGACCTGTTCGTCGCCGAACACCTCGATGAACTGACCGGCGCCCGCCATCTGCTGAGCAATGACCTCGGCGCCGCCTCGGCCCTCGCCTGGCGCCTGCGTCGTAGCGACGTGACGCTCTACGACACGCGCGGCGAGCTGAAGTACGGCCTCTCCTACCCCGAGCACAGCCAGCGCAGCGTGCCCTTGGCAGACATCCGCCAGTGGCTCTGGCGGGCACGCCAGGACGGCTCGATTGCCGTGCTGCTGCGGATCAACAGCGCCAGCGACCGCTACCAGCTGGCGCTGTTGCCGGGCGACGGCGAACGCTACCGGAACGGCAACCTGGTCCTGGCGATCCTGCCGCAGGTGCGCCCATGA |
| Protein Properties |
| Protein Residues: |
549 |
| Protein Molecular Weight: |
61.7 kDa |
| Protein Theoretical pI: |
10.1 |
| Hydropathicity (GRAVY score): |
0.399 |
| Charge at pH 7 (predicted): |
20.93 |
| Protein Sequence: |
>PA3556
MSRRQTCSLLLIAFGLFYLVPLSNHGLWIPDETRYAQISQAMLLGGDWVSPHFLGLRYFEKPVAGYWMIALGQAVFGENLFGVRIASVVATALSVLLAYLLARRLWRDPRTSLACALLYASFGLIAGQSGYANLDPQFTFWVNLSLVALWHALDAGSRRARLLGWTLLGLACGMGFLTKGFLAWLLPVLVALPYMLWQRRWRELLGYGALAVLAALLVCLPWALAVHAREADYWRFFFWHEHIRRFAGEDAQHSRPWWFYLPLLAVACLPWSGLLPSALRQAWHERRQAPVVFLALWLLLPLAFFSLSRGKLPTYIMPCLLPLALLMGHALVQRLRLGNSVALRGNGLLNLGLALLALAALAYLQLRKPVYQEEPFELFLVLLVIGAWAAAGLAQWRYPLRAWAAPLLASWVLIALLPAAMPNHVVQNKTPDLFVAEHLDELTGARHLLSNDLGAASALAWRLRRSDVTLYDTRGELKYGLSYPEHSQRSVPLADIRQWLWRARQDGSIAVLLRINSASDRYQLALLPGDGERYRNGNLVLAILPQVRP |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3556 and its homologs
|