|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001892 |
|---|
|
Identification |
|---|
| Name: |
4-Amino-4-deoxy-L-arabinose |
|---|
| Description: | 4-amino-4-deoxy-L-arabinose is invovled in Modification of lipid A. Attachment of the cationic sugar 4-amino-4-deoxy-l-arabinose (l-Ara4N) to lipid A is required for the maintenance of polymyxin resistance in Pseudomonas aeruginosa. The enzyme called AmT synthesizes this molecule and transfers it to lipid A. |
|---|
|
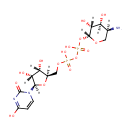
Structure |
|
|---|
| Synonyms: | - UDP-4-amino-4-Deoxy-b-L-arabinopyranose
- UDP-4-Amino-4-deoxy-beta-L-arabinopyranose
- UDP-4-Amino-4-deoxy-L-arabinose
- UDP-4-amino-4-Deoxy-β-L-arabinopyranose
- UDP-Ara4N
- UDP-L-Ara4N
|
|---|
|
Chemical Formula: |
C14H23N3O15P2 |
|---|
| Average Molecular Weight: |
535.291 |
|---|
| Monoisotopic Molecular
Weight: |
535.060440105 |
|---|
| InChI Key: |
GWBAKYBSWHQNMQ-IAZOVDBXSA-N |
|---|
| InChI: | InChI=1S/C14H23N3O15P2/c15-5-3-28-13(11(22)8(5)19)31-34(26,27)32-33(24,25)29-4-6-9(20)10(21)12(30-6)17-2-1-7(18)16-14(17)23/h1-2,5-6,8-13,19-22H,3-4,15H2,(H,24,25)(H,26,27)(H,16,18,23)/t5-,6+,8-,9+,10+,11+,12+,13+/m0/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | [({[(2R,3R,4S,5S)-5-amino-3,4-dihydroxyoxan-2-yl]oxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy})phosphinic acid |
|---|
|
Traditional IUPAC Name: |
{[(2R,3R,4S,5S)-5-amino-3,4-dihydroxyoxan-2-yl]oxy(hydroxy)phosphoryl}oxy[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxyphosphinic acid |
|---|
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(=O)O[C@@]2([H])OC[C@]([H])(N)[C@]([H])(O)[C@@]2([H])O)O[C@@]([H])(N2C=CC(O)=NC2=O)[C@]([H])(O)[C@]1([H])O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribobucleotides with diphosphate group linked to the ribose moiety. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
|
Class |
Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine ribonucleotides |
|---|
|
Direct Parent |
Pyrimidine ribonucleoside diphosphates |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Pyrimidine ribonucleoside diphosphate
- N-glycosyl compound
- Glycosyl compound
- Organic pyrophosphate
- Monosaccharide phosphate
- Hydroxypyrimidine
- Monoalkyl phosphate
- Pyrimidone
- Alkyl phosphate
- Pyrimidine
- Phosphoric acid ester
- Oxane
- Organic phosphoric acid derivative
- Organic phosphate
- Monosaccharide
- Hydropyrimidine
- Saccharide
- Heteroaromatic compound
- Oxolane
- Secondary alcohol
- 1,2-diol
- 1,2-aminoalcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Amine
- Alcohol
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aromatic heteromonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
Not Available |
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
| Resource | Link |
|---|
| CHEBI ID | 47025 | | HMDB ID | Not Available | | Pubchem Compound ID | 17756769 | | Kegg ID | C16153 | | ChemSpider ID | 21865443 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|