| Identification |
| Name: |
Adenylosuccinate synthetase |
| Synonyms: |
- AMPSase
- AdSS
- IMP--aspartate ligase
|
| Gene Name: |
purA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in adenylosuccinate synthase activity |
| Specific Function: |
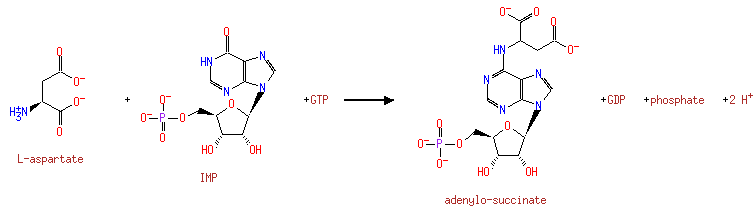
Plays an important role in the de novo pathway of purine nucleotide biosynthesis. Catalyzes the first commited step in the biosynthesis of AMP from IMP |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
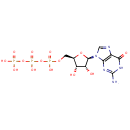 | + | 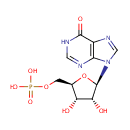 | + | L-Aspartic acid | + | 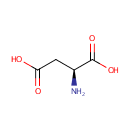 | → | 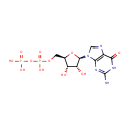 | + | 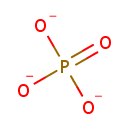 | + | N(6)-(1,2-dicarboxyethyl)AMP |
| | |
 | + | L-Aspartic acid | + |  | + |  | → |  | + |  | + | 2  | + | N(6)-(1,2-dicarboxyethyl)AMP | + | 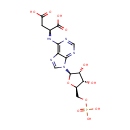 |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| adenylosuccinate synthase activity | | binding | | catalytic activity | | cation binding | | GTP binding | | guanyl nucleotide binding | | guanyl ribonucleotide binding | | ion binding | | ligase activity | | ligase activity, forming carbon-nitrogen bonds | | magnesium ion binding | | metal ion binding | | nucleotide binding | | purine nucleotide binding | | Process |
|---|
| cellular nitrogen compound metabolic process | | metabolic process | | nitrogen compound metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleoside phosphate metabolic process | | nucleotide metabolic process | | purine nucleotide biosynthetic process | | purine nucleotide metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4938 |
| Strand: |
- |
| Entrez Gene ID: |
877657 |
| Accession: |
NP_253625.1 |
| GI: |
15600131 |
| Sequence start: |
5542073 |
| Sequence End: |
5543365 |
| Sequence Length: |
1292 |
| Gene Sequence: |
>PA4938
ATGGGTAAGAATGTCGTAGTCCTGGGCACCCAATGGGGTGATGAGGGCAAGGGCAAGATCGTCGACCTGCTCACCGAGCAGGCGGCTGCCGTAGTTCGCTACCAGGGTGGTCACAACGCTGGCCACACCCTGGTGATCGATGGCGAGAAAACCGTCCTGCACCTGATTCCATCGGGCATCCTGCGCGAAGGCGTGCAGTGCCTGATCGGCAATGGCGTGGTGCTGGCGCCGGATGCGCTGCTGCGCGAAATCACCAAGCTGGAAGAGAAGGGCGTGCCGGTGCGCGAGCGCCTGCGCATCAGTCCTTCCTGTCCGCTGATCCTGTCCTATCACGTCGCCCTCGACCAGGCGCGCGAGAAGGCCCGCGGCGAAGCCAAGATCGGCACCACCGGTCGTGGCATCGGCCCGGCATACGAGGACAAGGTGGCACGTCGCGGCCTGCGCGTCGGCGACCTGTTCCACCGCGAGCGCTTCGCCGCCAAGCTGGGCGAGCTGCTGGACTACCATAACTTCGTCCTGCAGCACTACTACAAGGAGCCGGCCATCGACTTCCAGAAGACCCTGGACGAGGCGATGGAGTATGCCGAGCTGCTCAAGCCGATGATGGCCGATGTCGCTGCGACCCTGCACGACCTGCGCAAGCACGGCAAGGACATCATGTTCGAAGGCGCCCAGGGATCGCTGCTGGACATCGACCATGGCACCTACCCCTACGTGACCAGTTCCAACACCACCGCTGGCGGTACCGCCACCGGTTCCGGCTTCGGCCCGCTGTACCTGGACTACGTACTGGGTATCACCAAGGCCTATACCACTCGCGTCGGCTCCGGTCCGTTCCCGACCGAGCTGTTCGACGACGTCGGCGCCTACCTGGCCAAGCGCGGCCACGAGTTCGGTGCCACCACCGGTCGCGCTCGTCGCTGCGGCTGGTTCGATGCCGTGATCCTGCGCCGCGCCATCGAGATCAACAGCATCTCCGGCCTGTGCCTGACCAAGCTCGACGTGCTCGATGGCCTGGATGTGGTGCGCCTGTGCGTCGGCTACAAGAATGCCGATGGCGATGTGCTGGAAGCGCCGACCGATGCCGACAGCTACATCGGCCTGCAGCCGGTCTACGAGGAAATGCCCGGCTGGAGCGAGTCGACCGTGGGTGCGAAGACCCTGGAAGAGTTGCCGGCCAACGCCCGCGCCTACATCAAGCGCGTGGAAGAGCTGGTGGGTGCGCCGATCGATATCATCTCCACCGGTCCGGACCGCAACGAGACCATCATCCTGCGCCATCCATTCGCCTGA |
| Protein Properties |
| Protein Residues: |
430 |
| Protein Molecular Weight: |
46.8 kDa |
| Protein Theoretical pI: |
5.95 |
| Hydropathicity (GRAVY score): |
-0.146 |
| Charge at pH 7 (predicted): |
-7.25 |
| Protein Sequence: |
>PA4938
MGKNVVVLGTQWGDEGKGKIVDLLTEQAAAVVRYQGGHNAGHTLVIDGEKTVLHLIPSGILREGVQCLIGNGVVLAPDALLREITKLEEKGVPVRERLRISPSCPLILSYHVALDQAREKARGEAKIGTTGRGIGPAYEDKVARRGLRVGDLFHRERFAAKLGELLDYHNFVLQHYYKEPAIDFQKTLDEAMEYAELLKPMMADVAATLHDLRKHGKDIMFEGAQGSLLDIDHGTYPYVTSSNTTAGGTATGSGFGPLYLDYVLGITKAYTTRVGSGPFPTELFDDVGAYLAKRGHEFGATTGRARRCGWFDAVILRRAIEINSISGLCLTKLDVLDGLDVVRLCVGYKNADGDVLEAPTDADSYIGLQPVYEEMPGWSESTVGAKTLEELPANARAYIKRVEELVGAPIDIISTGPDRNETIILRHPFA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4938 and its homologs
|