| Identification |
| Name: |
UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase |
| Synonyms: |
- Polymyxin resistance protein pmrH
- UDP-(beta-L-threo-pentapyranosyl-4''-ulose diphosphate) aminotransferase
- UDP-Ara4O aminotransferase
|
| Gene Name: |
arnB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in transaminase activity |
| Specific Function: |
Catalyzes the conversion of UDP-4-keto-arabinose (UDP- Ara4O) to UDP-4-amino-4-deoxy-L-arabinose (UDP-L-Ara4N). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
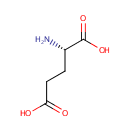
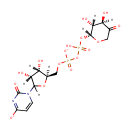
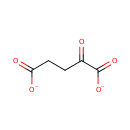
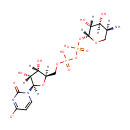
| PAMDB001892 | 4-Amino-4-deoxy-L-arabinose | MetaboCard | | PAMDB000466 | alpha-Ketoglutarate | MetaboCard | | PAMDB000055 | L-Glutamate | MetaboCard | | PAMDB000623 | Oxoglutaric acid | MetaboCard | | PAMDB001697 | UDP-4-Keto-pyranose | MetaboCard | | PAMDB003592 | UDP-beta-L-Threo-pentapyranos-4-ulose | MetaboCard | | PAMDB006357 | UDP-β-L-threo-pentapyranos-4-ulose | MetaboCard | | PAMDB001762 | Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | cofactor binding | | pyridoxal phosphate binding |
|
| Gene Properties |
| Locus tag: |
PA3552 |
| Strand: |
+ |
| Entrez Gene ID: |
879143 |
| Accession: |
NP_252242.1 |
| GI: |
15598748 |
| Sequence start: |
3979860 |
| Sequence End: |
3981008 |
| Sequence Length: |
1148 |
| Gene Sequence: |
>PA3552
ATGTCACTGGACTTTCTGCCATTCTCCCGGCCAAGCATCGGCGAGGACGAAATCGCCGCCGTGGAGCAGGTCCTGCGTTCCGGCTGGATCACCACCGGGCCGAAGAACCAGGAGCTCGAACAGCGCTTCGCCGAACGCCTCGGCTGCCGCCACGCGGTGGCGCTATCCTCGGCCACCGGCGCCCTGCACGTCACCCTGCTGGCGCTGGGCATCGGCCCCGGCGACGAAGTGATCACGCCGTCGCTGACCTGGGTCTCCACCGCCAACGTCATCACCCTGCTCGGCGCCACCCCGGTGTTCGTCGACGTCGACCGCGACACCCTGATGTGTTCGGCGCAGGCAGTGGAAGCCGCCATCGGCCCGCGCACCCGCGCCATCGTGCCGGTGCACTACGCCGGCTCCACCCTGGATCTCGAAGGCCTGCGCACGGTCGCCGGGCGCCACGGCATCGCCCTGGTCGAGGACGCCGCCCACGCGGTCGGCAGCGAATACCGCGGGCGCCCGGTAGGCTCCCGCGGCACGGCGATCTTTTCCTTCCACGCGATCAAGAACCTGACCTGCGCCGAAGGCGCGATGTTCGTCAGCGACGACAGTGCCCTCGCCGAACGGGTCCGCCGCCTCAAGTTCCACGGCCTCGGGGTCGATGCCTACGACCGCCTGAGCCATGGACGCAAGCCGCAGGCGGAAGTTATCGAGCCCGGTTTCAAATACAACCTGGCCGACCTCAACGCCGCCCTCGCGCTGGTCCAGCTCAAGCGCCTGGACGCGCTCAACGCGCGCCGCCAGGCCCTCGCCGAACGCTACCTGGAGCGCCTCGCCGGCCTGCCGCTGGCGCCGCTCGGCCTGCCCGCGCACAAGCAGCGCCACGCCTGGCACCTGTTCATCCTGCGCATCGACGCCGAGGTCTGCGGCCTGGGCCGCGACGCCTTCATGGAAGCCCTGAAGGCGCGCGGCATCGGTAGCGGCATCCATTTCATCGCCAGTCACCTGCACCACTACTACCGCCAGCGCCAGCCGCGCCTGAGCCTGCCCAACAGCGAGTGGAACTCGGCCCGCCTGTGCTCCATCCCGCTGTTCCCCGACATGCGCGACGACGACATCGAGCGCGTCGCCCGCGCCATCGAGGAGATCCTGGAGAAACGTCGATGA |
| Protein Properties |
| Protein Residues: |
382 |
| Protein Molecular Weight: |
41.9 kDa |
| Protein Theoretical pI: |
8 |
| Hydropathicity (GRAVY score): |
-0.061 |
| Charge at pH 7 (predicted): |
4.47 |
| Protein Sequence: |
>PA3552
MSLDFLPFSRPSIGEDEIAAVEQVLRSGWITTGPKNQELEQRFAERLGCRHAVALSSATGALHVTLLALGIGPGDEVITPSLTWVSTANVITLLGATPVFVDVDRDTLMCSAQAVEAAIGPRTRAIVPVHYAGSTLDLEGLRTVAGRHGIALVEDAAHAVGSEYRGRPVGSRGTAIFSFHAIKNLTCAEGAMFVSDDSALAERVRRLKFHGLGVDAYDRLSHGRKPQAEVIEPGFKYNLADLNAALALVQLKRLDALNARRQALAERYLERLAGLPLAPLGLPAHKQRHAWHLFILRIDAEVCGLGRDAFMEALKARGIGSGIHFIASHLHHYYRQRQPRLSLPNSEWNSARLCSIPLFPDMRDDDIERVARAIEEILEKRR |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3552 and its homologs
|