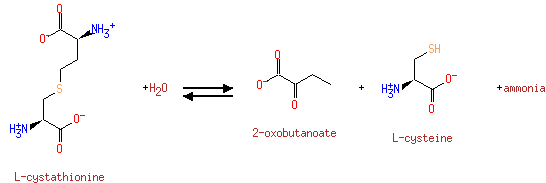
probable cystathionine gamma-lyase cystathionase; gamma-cystathionase; cysteine desulfhydrase; cystine desulfhydrase; homoserine dehydratase; homoserine deaminase (PA0400)
| Identification | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name: | probable cystathionine gamma-lyase cystathionase; gamma-cystathionase; cysteine desulfhydrase; cystine desulfhydrase; homoserine dehydratase; homoserine deaminase | ||||||||||||||||
| Synonyms: | metC; metB | ||||||||||||||||
| Gene Name: | Not Available | ||||||||||||||||
| Enzyme Class: | Not Available | ||||||||||||||||
| Biological Properties | |||||||||||||||||
| General Function: | Not Available | ||||||||||||||||
| Specific Function: | pyridoxal phosphate binding, catalytic activity | ||||||||||||||||
| Cellular Location: | Cytoplasmic | ||||||||||||||||
| KEGG Pathways: |
| ||||||||||||||||
| KEGG Reactions: | Not Available | ||||||||||||||||
| SMPDB Reactions: | Not Available | ||||||||||||||||
| PseudoCyc/BioCyc Reactions: |
| ||||||||||||||||
| Complex Reactions: | Not Available | ||||||||||||||||
| Transports: | Not Available | ||||||||||||||||
| Metabolites: | Not Available | ||||||||||||||||
| GO Classification: |
| ||||||||||||||||
| Gene Properties | |||||||||||||||||
| Locus tag: | PA0400 | ||||||||||||||||
| Strand: | + | ||||||||||||||||
| Entrez Gene ID: | 878286 | ||||||||||||||||
| Accession: | NP_249091.1 | ||||||||||||||||
| GI: | 15595597 | ||||||||||||||||
| Sequence start: | 442008 | ||||||||||||||||
| Sequence End: | 443192 | ||||||||||||||||
| Sequence Length: | 1184 | ||||||||||||||||
| Gene Sequence: |
>PA0400 ATGAGCCAGCACGACCAGCATCCCGACGCCCCGGCCCAGGCCTTTGCGACCCGGGTCATCCACGCCGGGCAGGCCCCCGATCCCTCGACCGGCGCGATCATGCCGCCGATCTACGCCAACTCCACCTATATCCAGGAAAGCCCCGGGGTACACAAGGGGCTCGACTACGGCCGCTCCCACAACCCGACCCGCTGGGCCCTGGAGCGCTGCGTGGCCGACCTCGAGGGCGGCACCCAGGCCTTCGCCTTCGCCTCCGGCCTGGCGGCGATCTCCTCGGTGCTGGAGCTGCTCGACGCCGGTTCCCACATCGTTTCCGGCAATGACCTCTACGGCGGTACCTTCCGCCTGTTCGAGCGGGTCCGCCGGCGCAGCGCCGGGCATCGCTTCAGCTTCGTCGACCCGACCGACCTGCAGGCCTTCGAGGCCGCGCTGACCCCGGAAACCCGCATGGTCTGGGTGGAAACCCCGAGCAACCCGCTGCTGCGCCTCACCGACCTGCGCGCCATCGCCCAGCTCTGCCGCGCCCGCGGCATCATCAGCGTGGCCGACAACACCTTCGCCAGCCCCTACATCCAGCGCCCCCTGGAGCTGGGCTTCGACGTGGTGGTGCACTCCACCACCAAGTACCTGAACGGCCATTCCGACGTGATCGGCGGCATCGCCATCGTCGGCGACAACCCGGACCTGCGCGAGCGCCTGGGCTTCCTGCAGAACTCGGTGGGCGCCATCTCCGGGCCGTTCGACGCCTTCCTCACCCTGCGCGGGGTGAAGACCCTGGCGCTGCGCATGGAGCGCCATTGCAGTAACGCCCTGGCCCTGGCCCAGTGGCTGGAGCGCCAGCCGCAAGTGGCGCGGGTCTACTACCCCGGCCTGGCTTCGCACCCGCAGCATGAGCTGGCGAAACGGCAGATGCGTGGCTTCGGCGGGATGATCTCCCTCGACCTGCGTTGCGACCTGGCCGGCGCCCGGCGCTTCCTGGAGAACGTGCGGATCTTCTCCCTGGCGGAGAGCCTGGGCGGGGTGGAAAGCCTGATCGAGCATCCGGCGATCATGACCCACGCCAGCATCCCGGCGGAAACCCGCGCCGACCTGGGCATTGGTGACTCGCTGATCCGCCTGTCGGTGGGCGTCGAGGCGCTGGAGGACCTGCAGGCCGACCTGGCGCAGGCGCTGGCGAAGATCTGA | ||||||||||||||||
| Protein Properties | |||||||||||||||||
| Protein Residues: | 394 | ||||||||||||||||
| Protein Molecular Weight: | 42.8 kDa | ||||||||||||||||
| Protein Theoretical pI: | 6.63 | ||||||||||||||||
| Hydropathicity (GRAVY score): | -0.053 | ||||||||||||||||
| Charge at pH 7 (predicted): | -2.75 | ||||||||||||||||
| Protein Sequence: |
>PA0400 MSQHDQHPDAPAQAFATRVIHAGQAPDPSTGAIMPPIYANSTYIQESPGVHKGLDYGRSHNPTRWALERCVADLEGGTQAFAFASGLAAISSVLELLDAGSHIVSGNDLYGGTFRLFERVRRRSAGHRFSFVDPTDLQAFEAALTPETRMVWVETPSNPLLRLTDLRAIAQLCRARGIISVADNTFASPYIQRPLELGFDVVVHSTTKYLNGHSDVIGGIAIVGDNPDLRERLGFLQNSVGAISGPFDAFLTLRGVKTLALRMERHCSNALALAQWLERQPQVARVYYPGLASHPQHELAKRQMRGFGGMISLDLRCDLAGARRFLENVRIFSLAESLGGVESLIEHPAIMTHASIPAETRADLGIGDSLIRLSVGVEALEDLQADLAQALAKI | ||||||||||||||||
| References | |||||||||||||||||
| External Links: |
| ||||||||||||||||
| General Reference: | PaperBLAST - Find papers about PA0400 and its homologs | ||||||||||||||||