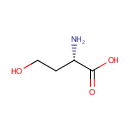
L-Homoserine (PAMDB000449)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000449 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Homoserine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Homoserine is a more reactive variant of the amino acid serine. In this variant, the hydroxyl side chain contains an additional CH2 group which brings the hydroxyl group closer to its own carboxyl group, allowing it to chemically react to form a five-membered ring. This occurs at the point that amino acids normally join to their neighbours in a peptide bond. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C4H9NO3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 119.1192 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 119.058243159 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | UKAUYVFTDYCKQA-VKHMYHEASA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C4H9NO3/c5-3(1-2-6)4(7)8/h3,6H,1-2,5H2,(H,7,8)/t3-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 672-15-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-4-hydroxybutanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-homoserine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CCO)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | L-alpha-amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 203 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Homoserine + NADP <> L-Aspartate-semialdehyde + Hydrogen ion + NADPH Adenosine triphosphate + L-Homoserine <> ADP + Hydrogen ion + O-Phosphohomoserine L-Homoserine + Succinyl-CoA <> Coenzyme A + O-Succinyl-L-homoserine Adenosine triphosphate + L-Homoserine <> ADP + O-Phosphohomoserine L-Homoserine + NAD <> L-Aspartate-semialdehyde + NADH + Hydrogen ion NAD(P)<sup>+</sup> + L-Homoserine < NAD(P)H + L-Aspartate-semialdehyde + Hydrogen ion L-Homoserine + NAD(P)(+) > L-Aspartate-semialdehyde + NAD(P)H L-Homoserine + Succinyl-CoA + L-Homoserine + Succinyl-CoA > Coenzyme A + O-Succinyl-L-homoserine L-Aspartate-semialdehyde + Hydrogen ion + NADPH + NADPH > NADP + L-Homoserine + L-Homoserine L-Homoserine + Adenosine triphosphate + L-Homoserine > Adenosine diphosphate + Hydrogen ion + O-Phosphohomoserine + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Kokusenya, Yoshio; Matsuoka, Manabu. Synthesis of amino acids by electrochemical reduction. I. Synthesis of L-homoserine by electrochemical reduction of L-asparagine. Denki Kagaku oyobi Kogyo Butsuri Kagaku (1987), 55(2), 174-5. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in ATP binding
- Specific function:
- Catalyzes the ATP-dependent phosphorylation of L- homoserine to L-homoserine phosphate. Is also able to phosphorylate the hydroxy group on gamma-carbon of L-homoserine analogs when the functional group at the alpha-position is a carboxyl, an ester, or even a hydroxymethyl group. Neither L- threonine nor L-serine are substrates of the enzyme
- Gene Name:
- thrB
- Locus Tag:
- PA5495
- Molecular weight:
- 35.4 kDa
Reactions
| ATP + L-homoserine = ADP + O-phospho-L-homoserine. |
Transporters
- General function:
- Involved in transport
- Specific function:
- Specific function unknown
- Gene Name:
- rhtA
- Locus Tag:
- PA1360
- Molecular weight:
- 30.7 kDa