| Identification |
| Name: |
Homoserine kinase |
| Synonyms: |
|
| Gene Name: |
thrB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in ATP binding |
| Specific Function: |
Catalyzes the ATP-dependent phosphorylation of L- homoserine to L-homoserine phosphate. Is also able to phosphorylate the hydroxy group on gamma-carbon of L-homoserine analogs when the functional group at the alpha-position is a carboxyl, an ester, or even a hydroxymethyl group. Neither L- threonine nor L-serine are substrates of the enzyme |
| Cellular Location: |
Cytoplasm (Probable) |
| KEGG Pathways: |
- Glycine, serine and threonine metabolism pae00260
- Metabolic pathways pae01100
- Microbial metabolism in diverse environments pae01120
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
L-Homoserine | + |  | + | 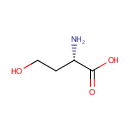 | → | Adenosine diphosphate | + |  | + | 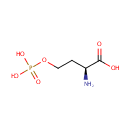 | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | catalytic activity | | homoserine kinase activity | | kinase activity | | nucleoside binding | | phosphotransferase activity, alcohol group as acceptor | | purine nucleoside binding | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| aspartate family amino acid metabolic process | | cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process | | cellular metabolic process | | metabolic process | | phosphate metabolic process | | phosphorus metabolic process | | phosphorylation | | threonine metabolic process |
|
| Gene Properties |
| Locus tag: |
PA5495 |
| Strand: |
+ |
| Entrez Gene ID: |
877825 |
| Accession: |
NP_254182.1 |
| GI: |
15600688 |
| Sequence start: |
6186927 |
| Sequence End: |
6187877 |
| Sequence Length: |
950 |
| Gene Sequence: |
>PA5495
ATGTCGGTGTTCACCCCACTCGAACGCTCGACCCTGGAAGCCTTCCTCGCCCCCTACGACCTGGGCCGCCTGCGGGATTTCCGTGGCATCGCCGAAGGCTCCGAGAACAGCAACTTCTTCGTCAGCCTGGAACACGGCGAGTTCGTCCTGACCCTGGTCGAGCGCGGCCCGGTGCAGGACCTGCCGTTCTTCATCGAACTGCTCGACGTGCTCCACGAGGACGGCCTGCCGGTGCCCTACGCCCTGCGCACCCGCGACGGCGAGGCCCTGCGCCGTCTCGAAGGCAAGCCGGCGCTGCTGCAACCGCGCCTGGCCGGCCGCCACGAGCGGCAGCCGAACGCCCACCACTGCCAGGAAGTGGGCGATCTGCTCGGCCACCTTCACGCCGCCACCCGCGGGCGCATCCTCGAACGCCCGAGCGACCGTGGCCTGCCCTGGATGCTGGAACAAGGTGCCAACCTCGCCCCGCGACTACCGGAACAGGCCCGCGCCCTGCTCGCCCCGGCCCTGGCGGAGATCGCCGCGCTGGATGCCGAGCGTCCAGCGCTGCCGCGCGCCAACCTGCATGCCGACCTGTTCCGCGACAACGTGCTGTTCGACGGCCCGCACCTGGCCGGCCTGATCGACTTCTACAACGCCTGCTCCGGCTGGATGCTCTACGACCTGGCGATCACCCTGAACGACTGGTGCTCCAATACCGACGGCAGCCTGGACCCGGCCCGCGCCCGCGCCCTGCTCGCCGCCTACGCCAACCGCCGGCCGTTCACCGCGCTGGAAGCCGAGCACTGGCCGAGCATGCTGCGGGTCGCCTGCGTGCGCTTCTGGCTGTCGCGACTGATCGCCGCCGAGGCGTTCGCCGGACAGGACGTGCTGATCCATGATCCGGCGGAGTTCGAGATACGCCTGGCGCAACGGCAGAACGTGGAGATTCATTTGCCGTTTGCGTTGTAG |
| Protein Properties |
| Protein Residues: |
316 |
| Protein Molecular Weight: |
35.4 kDa |
| Protein Theoretical pI: |
5.56 |
| Hydropathicity (GRAVY score): |
-0.109 |
| Charge at pH 7 (predicted): |
-9.22 |
| Protein Sequence: |
>PA5495
MSVFTPLERSTLEAFLAPYDLGRLRDFRGIAEGSENSNFFVSLEHGEFVLTLVERGPVQDLPFFIELLDVLHEDGLPVPYALRTRDGEALRRLEGKPALLQPRLAGRHERQPNAHHCQEVGDLLGHLHAATRGRILERPSDRGLPWMLEQGANLAPRLPEQARALLAPALAEIAALDAERPALPRANLHADLFRDNVLFDGPHLAGLIDFYNACSGWMLYDLAITLNDWCSNTDGSLDPARARALLAAYANRRPFTALEAEHWPSMLRVACVRFWLSRLIAAEAFAGQDVLIHDPAEFEIRLAQRQNVEIHLPFAL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5495 and its homologs
|





