Record Information Version
1.0 Update Date
1/22/2018 11:54:54 AM
Metabolite ID PAMDB110688
Identification Name:
2-aminoprop-2-enoate Description: Not Available
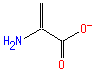
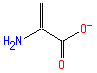
Structure
Synonyms:
2-aminoacrylate
α-aminoacrylate
2-aminoacrylic acid
2,3-didehydroalanine
α,β-dehydroalanine
anhydroserine 2-aminopropenoic acid
dehydroalanine
Chemical Formula:
C3 H4 NO2
Average Molecular Weight:
87.0320284099 Monoisotopic Molecular
Weight:
87.0320284099 InChI Key:
UQBOJOOOTLPNST-UHFFFAOYSA-M InChI:
InChI=1S/C3H5NO2/c1-2(4)3(5)6/h1,4H2,(H,5,6)/p-1 CAS
number:
Not Available IUPAC Name: Not Available
Traditional IUPAC Name:
Not Available SMILES: C=C(N)C(=O)[O-]
Chemical Taxonomy
Taxonomy Description This compound belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon).
Kingdom
Organic compounds Super Class Organic acids and derivatives
Class
Carboxylic acids and derivatives Sub Class Amino acids, peptides, and analogues
Direct Parent
Alpha amino acids Alternative Parents
Substituents
Alpha-amino acid Amino acid Carboxylic acid Enamine Monocarboxylic acid or derivatives Hydrocarbon derivative Primary amine Organooxygen compound Organonitrogen compound Amine Primary aliphatic amine Organic nitrogen compound Carbonyl group Organic oxygen compound Organic oxide Organopnictogen compound Organic anion Aliphatic acyclic compound Molecular Framework
Aliphatic acyclic compounds External Descriptors
alpha-amino-acid anion (CHEBI:58020) a small molecule (2-AMINOACRYLATE)
Physical Properties State:
Not Available Charge: Not Available
Melting point:
Not Available Experimental Properties:
Not Available Predicted Properties
Biological Properties Cellular Locations:
Not Available Reactions:
Pathways:
superpathway of sulfur amino acid biosynthesis (Saccharomyces cerevisiae )PWY-821 superpathway of L-lysine, L-threonine and L-methionine biosynthesis IP4-PWY L-methionine biosynthesis IHOMOSER-METSYN-PWY L-homocysteine and L-cysteine interconversionPWY-801 L-serine degradationSERDEG-PWY L-cysteine degradation IILCYSDEG-PWY glycine betaine degradation IPWY-3661
Spectra Spectra:
Not Available
References References:
Not Available Synthesis Reference:
Not Available Material Safety Data Sheet (MSDS)
Not Available
Links External Links:
This project is supported by
the University of Maryland ,
School of Pharmacy ,
Mass Spectrometry Center , a Waters Center of Excellence. The center is an NIH-investigator funded research and core facility that supports a wide range of cutting-edge metabolomic studies. The Center is supported through a center grant from the University of Maryland and NIH grants to its members. The PAMDB project is affiliated with
The Metabolomics Innovation Centre (TMIC) a leading metabolomics research and service center funded through Genome Alberta, Genome British Columbia and Genome Canada.