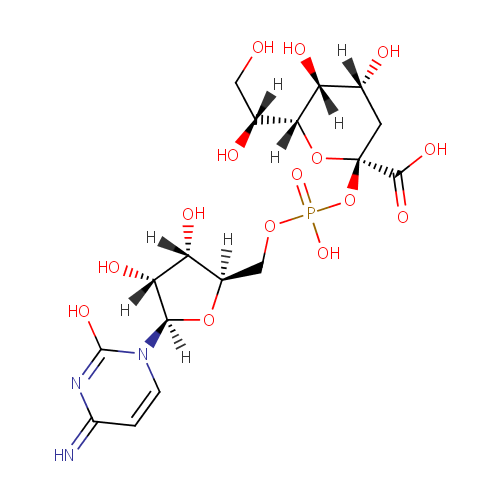
CMP-3-Deoxy-D-manno-octulosonate (PAMDB000981)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000981 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | CMP-3-Deoxy-D-manno-octulosonate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | CMP-3-deoxy-D-manno-octulosonate is a member of the chemical class known as Pyrimidine Nucleotide Sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. CMP KDO is involved in the biosynthesis of lipopolysaccharides. CKS is a pharmaceutical target because CMP-KDO is used in the biosynthesis of lipopolysaccharides that are vital for Gram-negative bacteria. (PMID 11545592) Pseudomonas aeruginosa KdtA (EcKdtA) is a bifunctional enzyme that transfers two KDO units from two CMP-KDO molecules to lipid IV(A). (PMID 20394418) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C17H26N3O15P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 543.3732 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 543.110153689 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | YWWJKULNWGRYAS-XKKDATLGSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C17H26N3O15P/c18-9-1-2-20(16(29)19-9)14-12(26)11(25)8(33-14)5-32-36(30,31)35-17(15(27)28)3-6(22)10(24)13(34-17)7(23)4-21/h1-2,6-8,10-14,21-26H,3-5H2,(H,27,28)(H,30,31)(H2,18,19,29)/t6-,7-,8-,10-,11-,12-,13-,14-,17+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S,4R,5R,6R)-2-[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(2-hydroxy-4-imino-1,4-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]-6-[(1R)-1,2-dihydroxyethyl]-4,5-dihydroxyoxane-2-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (2S,4R,5R,6R)-2-({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(2-hydroxy-4-iminopyrimidin-1-yl)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy)-6-[(1R)-1,2-dihydroxyethyl]-4,5-dihydroxyoxane-2-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@@](O)(CO)[C@@]1([H])O[C@@](C[C@@]([H])(O)[C@@]1([H])O)(OP(O)(=O)OC[C@@]1([H])O[C@@]([H])(N2C=CC(=N)N=C2O)[C@]([H])(O)[C@]1([H])O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as pyrimidine ribonucleoside monophosphates. These are pyrimidine ribobucleotides with monophosphate group linked to the ribose moiety. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrimidine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrimidine ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrimidine ribonucleoside monophosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Cytidine triphosphate + 3-Deoxy-D-manno-octulosonate <> CMP-3-Deoxy-D-manno-octulosonate + Pyrophosphate CMP-3-Deoxy-D-manno-octulosonate + Phospho-heptosyl-phospho-heptosyl-heptosyl-kdo2-lipidA > Cytidine monophosphate + Hydrogen ion + Kdo-phospho-heptosyl-phospho-heptosyl-heptosyl-kdo2-lipidA CMP-3-Deoxy-D-manno-octulosonate + 2,3,2'3'-Tetrakis(beta-hydroxymyristoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate > Cytidine monophosphate + Hydrogen ion + KDO-lipid IV(A) CMP-3-Deoxy-D-manno-octulosonate + KDO-lipid IV(A) > Cytidine monophosphate + Hydrogen ion + KDO(2)-lipid IV(A) 2,3,2'3'-Tetrakis(3-hydroxytetradecanoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate + CMP-3-Deoxy-D-manno-octulosonate + 2,3,2'3'-Tetrakis(beta-hydroxymyristoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate <> KDO-lipid IV(A) + Cytidine monophosphate KDO-lipid IV(A) + CMP-3-Deoxy-D-manno-octulosonate <> Di[3-deoxy-D-manno-octulosonyl]-lipid IV(A) + Cytidine monophosphate Di[3-deoxy-D-manno-octulosonyl]-lipid IV(A) + CMP-3-Deoxy-D-manno-octulosonate <> alpha-Kdo-(2->8)-alpha-Kdo-(2->4)-alpha-Kdo-(2->6)-lipid IVA + Cytidine monophosphate a lipopolysaccharide + CMP-3-Deoxy-D-manno-octulosonate KDOIII-lipopolysaccharide + Cytidine monophosphate CMP-3-Deoxy-D-manno-octulosonate + lipid IV<sub>A</sub> <> Hydrogen ion + KDO-lipid IV(A) + Cytidine monophosphate KDO-lipid IV(A) + CMP-3-Deoxy-D-manno-octulosonate <> Hydrogen ion + α-Kdo-(2->4)-α-Kdo-(2->6)-lipid IV<SUB>A</SUB> + Cytidine monophosphate 2,3,2'3'-Tetrakis(beta-hydroxymyristoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate + CMP-3-Deoxy-D-manno-octulosonate > alpha-Kdo-(2->6)-lipid IV(A) + Cytidine monophosphate Alpha-Kdo-(2->6)-lipid IV(A) + CMP-3-Deoxy-D-manno-octulosonate > alpha-Kdo-(2->4)-alpha-Kdo-(2->6)-lipid IV(A) + Cytidine monophosphate 3-deoxy-D-manno-octulosonate + Cytidine triphosphate + 3-Deoxy-D-manno-octulosonate > Pyrophosphate + CMP-3-Deoxy-D-manno-octulosonate CMP-3-Deoxy-D-manno-octulosonate + (2-N,3-O-bis(3-Hydroxytetradecanoyl)-4-O-phosphono-beta-D-glucosaminyl)-(1->6)-(2-N,3-O-bis(3-hydroxytetradecanoyl)-beta-D-glucosaminyl phosphate) > Cytidine monophosphate + Hydrogen ion + alpha-Kdo-(2??)-lipid IVA + Cytidine monophosphate alpha-Kdo-(2??)-lipid IVA + CMP-3-Deoxy-D-manno-octulosonate > Cytidine monophosphate + Hydrogen ion + a-Kdo-(2->4)-a-Kdo-(2->6)-lipid IVA + Cytidine monophosphate + a-Kdo-(2->4)-a-Kdo-(2->6)-lipid IVA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in lipopolysaccharide biosynthetic process
- Specific function:
- Activates KDO (a required 8-carbon sugar) for incorporation into bacterial lipopolysaccharide in Gram-negative bacteria
- Gene Name:
- kdsB
- Locus Tag:
- PA2979
- Molecular weight:
- 27.6 kDa
Reactions
| CTP + 3-deoxy-D-manno-octulosonate = diphosphate + CMP-3-deoxy-D-manno-octulosonate. |
- General function:
- Involved in biosynthetic process
- Specific function:
- Essential step in lipopolysaccharides biosynthesis. Acts at transfer of 3-deoxy-D-mono octulonic acid (KDO) from CMP-KDO to a tetraacyldisaccharide 1,4'-bisphosphate precursor of lipid A (lipid IVA). Transfers two molecules of KDO to lipid IVA. Degraded by FtsH; therefore FtsH regulates the addition of the sugar moiety of the LPS and thus the maturation of the LPS precursor
- Gene Name:
- waaA
- Locus Tag:
- PA4988
- Molecular weight:
- 46.5 kDa