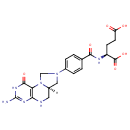
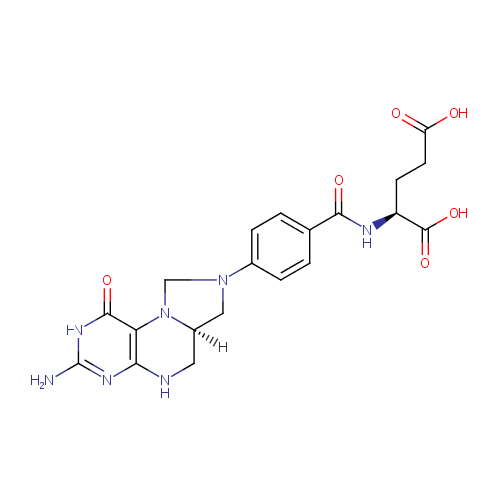
5,10-Methylene-THF (PAMDB000353)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000353 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 5,10-Methylene-THF | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 5,10-Methylene-THF is an intermediate in the metabolism of methane and the metabolism of nitrogen. 5,10-Methylenetetrahydrofolate (5,10-CH2-THF) is the substrate used by the enzyme methylenetetrahydrofolate reductase (MTHFR) to generate 5-methyltetrahydrofolate (5-MTHF, or levomefolic acid). 5,10-CH2-THF can also be used as a coenzyme in the biosynthesis of thymidine. More specifically it is the C1-donor in the reactions catalyzed by thymidylate synthase and thymidylate synthase (FAD). It also acts as a coenzyme in the synthesis of serine from glycine via the enzyme serine hydroxymethyl transferase. Methylenetetrahydrofolate reductase catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a cosubstrate for homocysteine remethylation to methionine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C20H23N7O6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 457.4399 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 457.170981503 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QYNUQALWYRSVHF-PZORYLMUSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C20H23N7O6/c21-20-24-16-15(18(31)25-20)27-9-26(8-12(27)7-22-16)11-3-1-10(2-4-11)17(30)23-13(19(32)33)5-6-14(28)29/h1-4,12-13H,5-9H2,(H,23,30)(H,28,29)(H,32,33)(H4,21,22,24,25,31)/t12-,13?/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 31690-11-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-({4-[(6aR)-3-amino-1-oxo-1H,2H,5H,6H,6aH,7H,8H,9H-imidazolidino[1,5-f]pteridin-8-yl]phenyl}formamido)pentanedioic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 5,10-Methylene-THF | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@@]12CN(CN1C1=C(NC2)N=C(N)NC1=O)C1=CC=C(C=C1)C(=O)NC(CCC(O)=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as hippuric acids. These are compounds containing hippuric acid, which consists of a of a benzoyl group linked to the N-terminal of a glycine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Benzenoids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Benzene and substituted derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Benzamides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hippuric acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Glycine + NAD + Tetrahydrofolic acid > Carbon dioxide + 5,10-Methylene-THF + NADH + Ammonium alpha-Ketoisovaleric acid + Water + 5,10-Methylene-THF + a-Ketoisovaleric acid <> 2-Dehydropantoate + Tetrahydrofolic acid 5,10-Methylene-THF + NADP <> 5,10-Methenyltetrahydrofolate + NADPH + Hydrogen ion L-Serine + Tetrahydrofolic acid <> Glycine + Water + 5,10-Methylene-THF dUMP + 5,10-Methylene-THF <> Dihydrofolic acid + 5-Thymidylic acid 2 Hydrogen ion + 5,10-Methylene-THF + NADH > 5-Methyltetrahydrofolic acid + NAD 5,10-Methylene-THF + NADP <> 5,10-Methenyltetrahydrofolate + NADPH Glycine + Tetrahydrofolic acid + NAD <> 5,10-Methylene-THF + Ammonia + Carbon dioxide + NADH + Hydrogen ion 5-Methyltetrahydrofolic acid + NADP <> 5,10-Methylene-THF + NADPH + Hydrogen ion S-Aminomethyldihydrolipoylprotein + Tetrahydrofolic acid + S-Aminomethyldihydrolipoylprotein <> Dihydrolipoylprotein + 5,10-Methylene-THF + Ammonia Formaldehyde + Tetrahydrofolic acid > 5,10-Methylene-THF + Water Water + alpha-Ketoisovaleric acid + 5,10-Methylene-THF <> 2-Dehydropantoate + Tetrahydrofolic acid [Protein]-S(8)-aminomethyldihydrolipoyllysine + Tetrahydrofolic acid > [protein]-dihydrolipoyllysine + 5,10-Methylene-THF + Ammonia 5-Methyltetrahydrofolic acid + NAD(P)(+) > 5,10-Methylene-THF + NAD(P)H 5,10-Methylene-THF + a-Ketoisovaleric acid + Water > Tetrahydrofolic acid + 2-Dehydropantoate More...5-Methyltetrahydrofolic acid + NAD + NADP <> 5,10-Methylene-THF + NADH + NADPH + Hydrogen ion Dihydrofolic acid + 5-Thymidylic acid + Dihydrofolic acid > 5,10-Methylene-THF + dUMP + 5,10-Methylene-THF 5,10-Methylene-THF + NADPH + Hydrogen ion + 5,10-Methylene-THF + NADPH > 5-Methyltetrahydrofolic acid + NADP + 5-Methyltetrahydrofolic acid S-Aminomethyldihydrolipoylprotein; + Tetrahydrofolic acid + Tetrahydrofolic acid <> 5,10-Methylene-THF + Ammonia + dihydrolipoylprotein + 5,10-Methylene-THF Tetrahydrofolic acid + L-Serine + Tetrahydrofolic acid + L-Serine <> 5,10-Methylene-THF + Glycine + Water + 5,10-Methylene-THF 5,10-Methenyltetrahydrofolic acid + NADPH + NADPH > 5,10-Methylene-THF + NADP + 5,10-Methylene-THF a-Ketoisovaleric acid + 5,10-Methylene-THF + Water + 5,10-Methylene-THF > Tetrahydrofolic acid + 2-dehydropantoate + Tetrahydrofolic acid + 2-Dehydropantoate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Agrawal, Nitish; Mihai, Cornelia; Kohen, Amnon. Microscale synthesis of isotopically labeled R-[6-xH]N5,N10-methylene-5,6,7,8-tetrahydrofolate as a cofactor for thymidylate synthase. Analytical Biochemistry (2004), 328(1), 44-50. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Interconversion of serine and glycine
- Gene Name:
- glyA
- Locus Tag:
- PA4602
- Molecular weight:
- 45.2 kDa
Reactions
| 5,10-methylenetetrahydrofolate + glycine + H(2)O = tetrahydrofolate + L-serine. |
- General function:
- Involved in thymidylate synthase activity
- Specific function:
- Provides the sole de novo source of dTMP for DNA biosynthesis. This protein also binds to its mRNA thus repressing its own translation
- Gene Name:
- thyA
- Locus Tag:
- PA0342
- Molecular weight:
- 30 kDa
Reactions
| 5,10-methylenetetrahydrofolate + dUMP = dihydrofolate + dTMP. |
- General function:
- Involved in methylenetetrahydrofolate reductase (NADPH) activity
- Specific function:
- 5-methyltetrahydrofolate + NAD(P)(+) = 5,10- methylenetetrahydrofolate + NAD(P)H
- Gene Name:
- metF
- Locus Tag:
- PA0430
- Molecular weight:
- 32.2 kDa
Reactions
| 5-methyltetrahydrofolate + NAD(P)(+) = 5,10-methylenetetrahydrofolate + NAD(P)H. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the oxidation of 5,10- methylenetetrahydrofolate to 5,10-methenyltetrahydrofolate and then the hydrolysis of 5,10-methenyltetrahydrofolate to 10- formyltetrahydrofolate. This enzyme is specific for NADP
- Gene Name:
- folD
- Locus Tag:
- PA1796
- Molecular weight:
- 30.5 kDa
Reactions
| 5,10-methylenetetrahydrofolate + NADP(+) = 5,10-methenyltetrahydrofolate + NADPH. |
| 5,10-methenyltetrahydrofolate + H(2)O = 10-formyltetrahydrofolate. |
- General function:
- Involved in aminomethyltransferase activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine
- Gene Name:
- gcvT
- Locus Tag:
- PA5215
- Molecular weight:
- 38.9 kDa
Reactions
| [Protein]-S(8)-aminomethyldihydrolipoyllysine + tetrahydrofolate = [protein]-dihydrolipoyllysine + 5,10-methylenetetrahydrofolate + NH(3). |
- General function:
- Involved in 3-methyl-2-oxobutanoate hydroxymethyltransferase activity
- Specific function:
- Catalyzes the reversible reaction in which hydroxymethyl group from 5,10-methylenetetrahydrofolate is tranferred onto alpha-ketoisovalerate to form ketopantoate
- Gene Name:
- panB
- Locus Tag:
- PA4729
- Molecular weight:
- 27.9 kDa
Reactions
| 5,10-methylenetetrahydrofolate + 3-methyl-2-oxobutanoate + H(2)O = tetrahydrofolate + 2-dehydropantoate. |
- General function:
- Involved in glycine dehydrogenase (decarboxylating) activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The P protein binds the alpha-amino group of glycine through its pyridoxal phosphate cofactor; CO(2) is released and the remaining methylamine moiety is then transferred to the lipoamide cofactor of the H protein
- Gene Name:
- gcvP
- Locus Tag:
- PA5213
- Molecular weight:
- 104.7 kDa
Reactions
| Glycine + H-protein-lipoyllysine = H-protein-S-aminomethyldihydrolipoyllysine + CO(2). |
- General function:
- Amino acid transport and metabolism
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The H protein shuttles the methylamine group of glycine from the P protein to the T protein
- Gene Name:
- gcvH
- Locus Tag:
- PA5214
- Molecular weight:
- 13.6 kDa