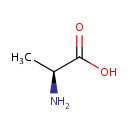
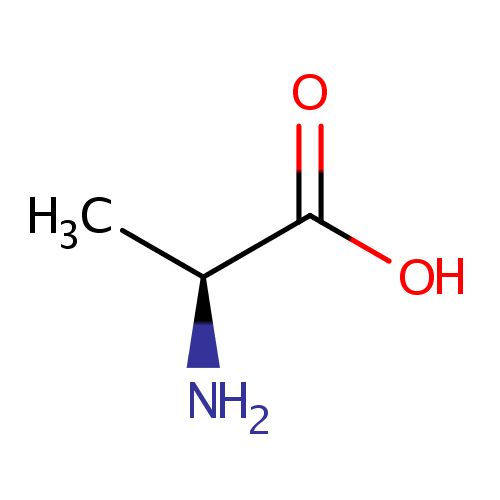
L-Alanine (PAMDB000063)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000063 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Alanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Alanine is an amino acid made from the conversion of the carbohydrate pyruvate or the breakdown of DNA and the dipeptides carnosine and anserine. Alanine is an important participant as well as regulator in glucose metabolism. Normal alanine metabolism, like that of other amino acids, is highly dependent upon enzymes that contain vitamin B6. (http://www.dcnutrition.com/AminoAcids/) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H7NO2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 89.0932 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 89.047678473 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QNAYBMKLOCPYGJ-REOHCLBHSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6)/t2-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 56-41-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-aminopropanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-alanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[C@H](N)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | L-alpha-amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 300 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Alanine + Pyridoxal 5'-phosphate > Pyridoxamine 5'-phosphate + Pyruvic acid L-Alanine <> D-Alanine L-Cysteine + SufSE sulfur acceptor complex > L-Alanine + SufSE with bound sulfur alpha-Ketoglutarate + L-Alanine <> L-Glutamate + Pyruvic acid Adenosine triphosphate + Water + L-Alanine > ADP + L-Alanine + Hydrogen ion + Phosphate Adenosine triphosphate + Water + L-Alanine > ADP + L-Alanine + Hydrogen ion + Phosphate L-Alanine + Adenosine triphosphate + UDP-N-Acetylmuraminate <> ADP + Hydrogen ion + Phosphate + UDP-N-Acetylmuramoyl-L-alanine L-Alanine-L-glutamate + Water > L-Alanine + L-Glutamate L-Alanine + Pimeloyl-[acyl-carrier protein] > 8-Amino-7-oxononanoate + acyl carrier protein + Carbon dioxide L-Cysteine + IscS sulfur acceptor protein > L-Alanine + IscS with bound sulfur L-Alanine + Adenosine triphosphate + tRNA(Ala) > L-Alanyl-tRNA(Ala) + Adenosine monophosphate + Pyrophosphate 3-Sulfinoalanine + 2 Hydrogen ion > L-Alanine + Sulfur dioxide alpha-Ketoisovaleric acid + L-Alanine <> Pyruvic acid + L-Valine + a-Ketoisovaleric acid L-Valine + Pyruvic acid <> alpha-Ketoisovaleric acid + L-Alanine Adenosine triphosphate + L-Alanine + tRNA(Ala) + tRNA(Ala) <> Adenosine monophosphate + Pyrophosphate + L-Alanyl-tRNA + L-Alanyl-tRNA Adenosine triphosphate + UDP-N-Acetylmuraminate + L-Alanine <> ADP + Phosphate + UDP-N-Acetylmuramoyl-L-alanine 6-Carboxyhexanoyl-CoA + L-Alanine + Pimeloyl-[acyl-carrier protein] <> 8-Amino-7-oxononanoate + Coenzyme A + Carbon dioxide + Acyl-carrier protein Selenocysteine + Reduced acceptor <> Hydrogen selenide + L-Alanine + Acceptor N-Acetylmuramoyl-Ala + Water <> N-Acetyl-D-muramoate + L-Alanine [Enzyme]-cysteine + L-Cysteine <> [Enzyme]-S-sulfanylcysteine + L-Alanine UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate-D-alanine + Water > L-Alanine + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate Hydrogen ion + L-Alanine + pimeloyl-CoA > Carbon dioxide + Coenzyme A + 8-Amino-7-oxononanoate Oxoglutaric acid + L-Alanine <> L-Glutamate + Pyruvic acid L-Alanine + Hydrogen ion + Pimeloyl-ACPs > 8-Amino-7-oxononanoate + Carbon dioxide + ACP More...L-Cysteine + a sulfur acceptor + Hydrogen ion L-Alanine + <i>S</i>-sulfanyl-[acceptor] 3-Sulfinoalanine + Water Hydrogen ion + L-Alanine + Sulfite L-Cysteine + L-Cysteine-Desulfurases > L-Alanine + Persulfurated-L-cysteine-desulfurases ala-asp + Water > L-Alanine + L-Aspartic acid ala-gln + Water > L-Alanine + L-Glutamine ala-gly + Water > L-Alanine + Glycine ala-his + Water > L-Alanine + L-Histidine ala-leu + Water > L-Alanine + L-Leucine ala-thr + Water > L-Alanine + L-Threonine L-Alanyl-L-Glutamate + Water > L-Alanine + L-Glutamate methionine-alanine dipeptide + Water > L-Methionine + L-Alanine a reduced electron acceptor + Selenocysteine <> L-Alanine + Selenium + an oxidized electron acceptor + Hydrogen ion L-Valine + Pyruvic acid > a-Ketoisovaleric acid + L-Alanine L-Cysteine + acceptor > L-Alanine + S-sulfanyl-acceptor Adenosine triphosphate + UDP-N-Acetylmuraminate + L-Alanine > ADP + Inorganic phosphate + UDP-N-Acetylmuramoyl-L-alanine a pimeloyl-[acp] + L-Alanine + L-Alanine > Carbon dioxide + a holo-[acyl-carrier protein] + 8-Amino-7-oxononanoate a sulfurated [sulfur carrier] + L-Alanine + L-Alanine > 8-Amino-7-oxononanoate + Coenzyme A + Carbon dioxide L-Alanine + L-Alanine <> D-Alanine UDP-N-acetylmuraminate + Adenosine triphosphate + L-Alanine + UDP-N-Acetylmuraminate + L-Alanine > UDP-N-Acetylmuramoyl-L-alanine + Adenosine diphosphate + Phosphate + ADP Adenosine triphosphate + L-Alanine + tRNA(Ala) + L-Alanine > Adenosine monophosphate + Pyrophosphate + L-alanyl-tRNA(Ala) L-Alanine + Oxoglutaric acid + L-Alanine <> L-Glutamic acid + Pyruvic acid + L-Glutamate L-Alanine + Glyoxylic acid + L-Alanine <> Glycine + Pyruvic acid L-Cysteine + an [L-cysteine desulfurase] L-cysteine persulfide > an [L-cysteine desulfurase] L-cysteine persulfide + L-Alanine + L-Alanine L-Valine + Pyruvic acid + L-Valine > L-Alanine + a-Ketoisovaleric acid + L-Alanine L-Alanine + Adenosine triphosphate + Hydrogen ion + tRNA(Ala) + L-Alanine > Pyrophosphate + Adenosine monophosphate + L-alanyl-tRNA(Ala) UDP-N-acetyl-α-D-muramate + L-Alanine + Adenosine triphosphate + L-Alanine > Adenosine diphosphate + Phosphate + Hydrogen ion + UDP-N-Acetylmuramoyl-L-alanine + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Chibata I; Kakimoto T; Kato J Enzymatic production of L-alanine by Pseudomonas dacunhae. Applied microbiology (1965), 13(5), 638-45. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- Edits incorrectly charged Ser-tRNA(Ala) and Gly- tRNA(Ala) but not incorrectly charged Ser-tRNA(Thr)
- Gene Name:
- alaS
- Locus Tag:
- PA0903
- Molecular weight:
- 94.7 kDa
Reactions
| ATP + L-alanine + tRNA(Ala) = AMP + diphosphate + L-alanyl-tRNA(Ala). |
- General function:
- Involved in alanine racemase activity
- Specific function:
- Provides the D-alanine required for cell wall biosynthesis
- Gene Name:
- alr
- Locus Tag:
- PA4930
- Molecular weight:
- 38.3 kDa
Reactions
| L-alanine = D-alanine. |
- General function:
- Involved in metabolic process
- Specific function:
- Catalyzes the removal of elemental sulfur and selenium atoms from cysteine and selenocysteine to produce alanine. Functions as a sulfur delivery protein for NAD, biotin and Fe-S cluster synthesis. Transfers sulfur on 'Cys-456' of thiI in a transpersulfidation reaction. Transfers sulfur on 'Cys-19' of tusA in a transpersulfidation reaction. Functions also as a selenium delivery protein in the pathway for the biosynthesis of selenophosphate
- Gene Name:
- iscS
- Locus Tag:
- PA3814
- Molecular weight:
- 44.7 kDa
Reactions
| L-cysteine + acceptor = L-alanine + S-sulfanyl-acceptor. |
- General function:
- Involved in catalytic activity
- Specific function:
- Interconversion of serine and glycine
- Gene Name:
- glyA
- Locus Tag:
- PA4602
- Molecular weight:
- 45.2 kDa
Reactions
| 5,10-methylenetetrahydrofolate + glycine + H(2)O = tetrahydrofolate + L-serine. |
- General function:
- Involved in 8-amino-7-oxononanoate synthase activity
- Specific function:
- Catalyzes the decarboxylative condensation of pimeloyl- CoA and L-alanine to produce 8-amino-7-oxononanoate (AON), coenzyme A, and carbon dioxide
- Gene Name:
- bioF
- Locus Tag:
- PA0501
- Molecular weight:
- 42.2 kDa
Reactions
| 6-carboxyhexanoyl-CoA + L-alanine = 8-amino-7-oxononanoate + CoA + CO(2). |
- General function:
- Involved in ATP binding
- Specific function:
- Cell wall formation
- Gene Name:
- murC
- Locus Tag:
- PA4411
- Molecular weight:
- 51.9 kDa
Reactions
| ATP + UDP-N-acetylmuramate + L-alanine = ADP + phosphate + UDP-N-acetylmuramoyl-L-alanine. |
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Cell-wall hydrolase probably involved in cell-wall hydrolysis, septation or recycling
- Gene Name:
- amiB
- Locus Tag:
- PA4947
- Molecular weight:
- 50.7 kDa
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
- General function:
- Involved in alanine racemase activity
- Specific function:
- Isomerizes L-alanine to D-alanine which is then oxidized to pyruvate by dadA
- Gene Name:
- dadX
- Locus Tag:
- PA5302
- Molecular weight:
- 38.9 kDa
Reactions
| L-alanine = D-alanine. |
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides
- Gene Name:
- amiA
- Locus Tag:
- PA5538
- Molecular weight:
- 42.9 kDa
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Cell-wall hydrolase probably involved in cell-wall hydrolysis, septation or recycling
- Gene Name:
- amiC
- Locus Tag:
- PA3364
- Molecular weight:
- 42.8 kDa
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
- General function:
- Involved in lyase activity
- Specific function:
- Catalyzes the cleavage of L-allo-threonine and L- threonine to glycine and acetaldehyde. L-threo-phenylserine and L- erythro-phenylserine are also good substrates
- Gene Name:
- ltaE
- Locus Tag:
- PA0902
- Molecular weight:
- 35.4 kDa
Reactions
| L-threonine = glycine + acetaldehyde. |
| L-allo-threonine = glycine + acetaldehyde. |
- General function:
- Defense mechanisms
- Specific function:
- Releases the terminal D-alanine residue from the cytoplasmic tetrapeptide recycling product L-Ala-gamma-D-Glu-meso- Dap-D-Ala. To a lesser extent, can also cleave D-Ala from murein derivatives containing the tetrapeptide, i.e. MurNAc-tetrapeptide, UDP-MurNAc-tetrapeptide, GlcNAc-MurNAc-tetrapeptide, and GlcNAc- anhMurNAc-tetrapeptide. Does not act on murein sacculi or cross- linked muropeptides. The tripeptides produced by the lcdA reaction can then be reused as peptidoglycan building blocks; lcdA is thereby involved in murein recycling. Is also essential for viability during stationary phase
- Gene Name:
- ldcA
- Locus Tag:
- PA1818
- Molecular weight:
- 82.8 kDa
Reactions
| GlcNAc-MurNAc-L-alanyl-gamma-D-glutamyl-meso-diaminopimelyl-D-alanine + H(2)O = GlcNAc-MurNAc-L-alanyl-gamma-D-glutamyl-meso-diaminopimelate + D-alanine. |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livH
- Locus Tag:
- PA1073
- Molecular weight:
- 32.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livM
- Locus Tag:
- PA1072
- Molecular weight:
- 45.6 kDa
- General function:
- Involved in 1-aminocyclopropane-1-carboxylate synthase activity
- Specific function:
- Specific function unknown
- Gene Name:
- yfbQ
- Locus Tag:
- PA2828
- Molecular weight:
- 44.8 kDa
Reactions
| L-alanine + 2-oxoglutarate = pyruvate + L-glutamate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livG
- Locus Tag:
- PA1071
- Molecular weight:
- 28.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livF
- Locus Tag:
- PA1070
- Molecular weight:
- 25.6 kDa
- General function:
- Amino acid transport and metabolism
- Specific function:
- Specific function unknown
- Gene Name:
- yfdZ
- Locus Tag:
- PA4715
- Molecular weight:
- 46.1 kDa
Reactions
| L-alanine + 2-oxoglutarate = pyruvate + L-glutamate. |
Transporters
- General function:
- Involved in sodium:amino acid symporter activity
- Specific function:
- Specific function unknown
- Gene Name:
- yaaJ
- Locus Tag:
- PA3641
- Molecular weight:
- 50.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Probably part of a binding-protein-dependent transport system yecCS for an amino acid. Probably responsible for energy coupling to the transport system
- Gene Name:
- yecC
- Locus Tag:
- PA5152
- Molecular weight:
- 28.4 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livH
- Locus Tag:
- PA1073
- Molecular weight:
- 32.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livM
- Locus Tag:
- PA1072
- Molecular weight:
- 45.6 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livG
- Locus Tag:
- PA1071
- Molecular weight:
- 28.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livF
- Locus Tag:
- PA1070
- Molecular weight:
- 25.6 kDa