Search Results for compounds
Searching compounds for
returned 4373 results.
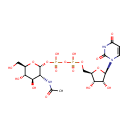
Uridine diphosphate-N-acetylglucosamine (PAMDB000120)
IUPAC:
[({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid
CAS: 528-04-1
Description: Uridine diphosphate N-acetylglucosamine or UDP-GlcNAc is a nucleotide sugar and a coenzyme in metabolism. It is used by glycosyltransferases to transfer N-acetylglucosamine residues to substrates. D-Glucosamine is made naturally in the form of glucosamine-6-phosphate, and is the biochemical precursor of all nitrogen-containing sugars. Specifically, glucosamine-6-phosphate is synthesized from fructose 6-phosphate and glutamine as the first step of the hexosamine biosynthesis pathway. The end-product of this pathway is UDP-GlcNAc, which is then used for making glycosaminoglycans, proteoglycans, and glycolipids.
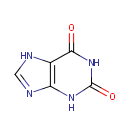
Xanthine (PAMDB000121)
IUPAC:
2,3,6,7-tetrahydro-1H-purine-2,6-dione
CAS: 69-89-6
Description: Xanthine is an intermediate in the degradation of adenosine monophosphate to uric acid, being formed by oxidation of hypoxanthine. The methylated xanthine compounds caffeine, theobromine, and theophylline and their derivatives are used in medicine for their bronchodilator effects. (Dorland, 28th ed.)
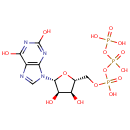
Xanthosine 5-triphosphate (PAMDB000122)
IUPAC:
({[({[(2R,3S,4R,5R)-5-(2,6-dihydroxy-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphonic acid
CAS: 6253-56-1
Description: Xanthosine 5-triphosphate (XTP) is a Guanosine triphosphate (GTP) analogue. The base of XTP, xanthine, bears a keto group instead of an amino group at C2 of the purine rings. XTP can substitute for GTP in supporting receptor-mediated adenylyl cyclase activation. Xanthosine 5-triphosphate is an intermediate of the Purine metabolism pathway, a substrate of the enzymes dinucleoside tetraphosphatase (EC 3.6.1.17) and nucleoside-triphosphate pyrophosphatase (EC 3.6.1.19). It is also a substrate for Xanthine triphosphatase, which is a phosphatase that hydrolyzes non-canonical purine nucleotides such as XTP and ITP to their respective diphosphate derivatives.
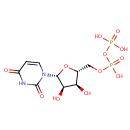
Uridine 5'-diphosphate (PAMDB000123)
IUPAC:
[({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphonic acid
CAS: 58-98-0
Description: Uridine 5'-diphosphate, also known as UDP, is an uracil nucleotide containing a pyrophosphate group esterified to C5 of the sugar moiety. UDP is an important extracellular pyrimidine signaling molecule that mediates diverse biological effects via P1 and P2 purinergic receptors, such as the uptake of thymidine and proliferation of gliomas. UDP induces intracellular Ca(2+) responses and oscillations in HeLa cells, due to the activation of P2Ys (G-protein coupled ATP receptors).
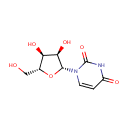
Uridine (PAMDB000124)
IUPAC:
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,3,4-tetrahydropyrimidine-2,4-dione
CAS: 58-96-8
Description: Uridine is a molecule (known as a nucleoside) that is formed when uracil is attached to a ribose ring (also known as a ribofuranose) via a b-N1-glycosidic bond. (Wikipedia)
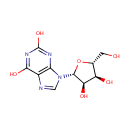
Xanthosine (PAMDB000125)
IUPAC:
9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-9H-purine-2,6-diol
CAS: 146-80-5
Description: Xanthosine is a nucleoside derived from xanthine and ribose. It is a deamination product of guanosine. Xanthosine monophosphate is an intermediate in purine metabolism, formed from IMP, and forming GMP.(Wikipedia)
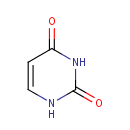
Uracil (PAMDB000126)
IUPAC:
1,2,3,4-tetrahydropyrimidine-2,4-dione
CAS: 66-22-8
Description: Uracil is a common naturally occurring pyrimidine found in RNA, it base pairs with adenine and is replaced by thymine in DNA. Methylation of uracil produces thymine. Uracil serves as allosteric regulator and coenzyme for many important biochemical reactions. Uracil is also involved in the biosynthesis of polysaccharides and the transportation of sugars containing aldehydes. In Pseudomonas aeruginosa, uracil catabolism is regulated by the amount of metabolically available nitrogen.
Uridine diphosphategalactose (PAMDB000127)
IUPAC:
[({[(2S,3R,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R,6S)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid
CAS: 2956-16-3
Description: Uridine diphosphategalactose (UDPgal) is a nucleoside diphosphate sugar which can be epimerized into UDPglucose for entry into the mainstream of carbohydrate metabolism. UDPgal is a pivotal compound in the metabolism of galactose. UDPgal is a product of the galactose-L-phosphate uridyl transferase (EC 2.7.7.10) reaction but may also be made from Glucose-L-P, involving uridine diphosphate galactose-4-epimerase (EC 5.1.3.2). UDPgal is the necessary galactosyl donor of galactose in the metabolism to incorporate it into complex oligosaccharides, glycoproteins and glycolipids (galactosides). (PMID: 2122114, 7671968)
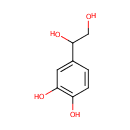
3,4-Dihydroxyphenylglycol (PAMDB000131)
IUPAC:
4-(1,2-dihydroxyethyl)benzene-1,2-diol
CAS: 28822-73-3
Description: (S)-3,5-Dihydroxyphenylglycine or DHPG is a potent agonist of group metabotropic glutamate receptors (mGluRs). It is an intermediate in tyrosine metabolism.
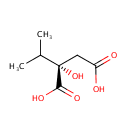
2-Isopropylmalic acid (PAMDB000135)
IUPAC:
(2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid
CAS: 3237-44-3
Description: 2-Isopropylmalic acid is involved in valine, leucine and isoleucine biosynthesis and pyruvate metabolism pathways. In pyruvate metabolism, 2-isopropylmalate synthase (EC:2.3.3.13) catalyzes the formation of 2-Isopropylmalic acid from acetyl-CoA. (KEGG)