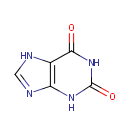
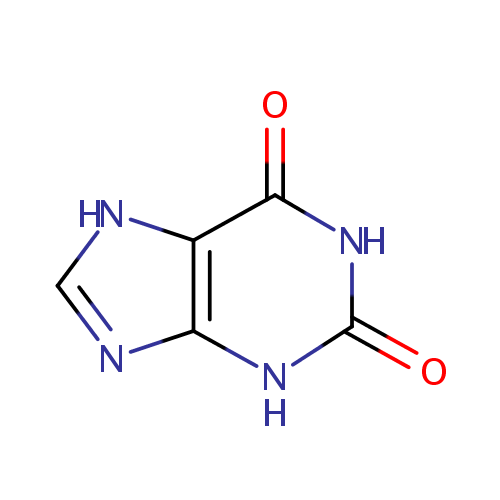
Xanthine (PAMDB000121)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000121 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Xanthine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Xanthine is an intermediate in the degradation of adenosine monophosphate to uric acid, being formed by oxidation of hypoxanthine. The methylated xanthine compounds caffeine, theobromine, and theophylline and their derivatives are used in medicine for their bronchodilator effects. (Dorland, 28th ed.) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H4N4O2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 152.1109 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 152.033425392 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | LRFVTYWOQMYALW-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H4N4O2/c10-4-2-3(7-1-6-2)8-5(11)9-4/h1H,(H3,6,7,8,9,10,11) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 69-89-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2,3,6,7-tetrahydro-1H-purine-2,6-dione | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | xanthine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O=C1NC2=C(NC=N2)C(=O)N1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as alkaloids and derivatives. These are naturally occurring chemical compounds that contain mostly basic nitrogen atoms. This group also includes some related compounds with neutral and even weakly acidic properties. Also some synthetic compounds of similar structure are attributed to alkaloids. In addition to carbon, hydrogen and nitrogen, alkaloids may also contain oxygen, sulfur and more rarely other elements such as chlorine, bromine, and phosphorus. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Alkaloids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alkaloids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | >300 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Water + Hypoxanthine + NAD > Hydrogen ion + NADH + Xanthine Water + NAD + Xanthine <> Hydrogen ion + NADH + Uric acid Water + Xanthosine > Ribose + Xanthine Phosphoribosyl pyrophosphate + Xanthine <> Pyrophosphate + Xanthylic acid Phosphate + Xanthosine <> Ribose-1-phosphate + Xanthine Guanine + Hydrogen ion + Water > Ammonium + Xanthine Guanine + Water <> Xanthine + Ammonia Xanthosine + Phosphate <> Xanthine + alpha-D-Ribose 1-phosphate isoguanine + Water > Xanthine + Ammonia Xanthosine + Water > D-ribose + Xanthine Xanthine + NAD + Water > Uric acid + NADH Hypoxanthine + NAD + Water > Xanthine + NADH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Procedure for the production of xanthine and xanthine-like materials. Fr. (1967), 4 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in hydrolase activity
- Specific function:
- Catalyzes the hydrolytic deamination of guanine, producing xanthine and ammonia
- Gene Name:
- guaD
- Locus Tag:
- PA1521
- Molecular weight:
- 48.3 kDa
Reactions
| Guanine + H(2)O = xanthine + NH(3). |
- General function:
- Involved in flavin adenine dinucleotide binding
- Specific function:
- Xanthine + NAD(+) + H(2)O = urate + NADH
- Gene Name:
- yagS
- Locus Tag:
- PA1932
- Molecular weight:
- 35.6 kDa
Reactions
| Xanthine + NAD(+) + H(2)O = urate + NADH. |
| Hypoxanthine + NAD(+) + H(2)O = xanthine + NADH. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Presumed to be a dehydrogenase, but possibly an oxidase. Participates in limited purine salvage (requires aspartate) but does not support aerobic growth on purines as the sole carbon source (purine catabolism). Deletion results in increased adenine sensitivity, suggesting that this protein contributes to the conversion of adenine to guanine nucleotides during purine salvage
- Gene Name:
- xdhA
- Locus Tag:
- PA1524
- Molecular weight:
- 52.9 kDa
Reactions
| Xanthine + NAD(+) + H(2)O = urate + NADH. |
| Hypoxanthine + NAD(+) + H(2)O = xanthine + NADH. |
- General function:
- Involved in flavin adenine dinucleotide binding
- Specific function:
- Presumed to be a dehydrogenase, but possibly an oxidase. Participates in limited purine salvage (requires aspartate) but does not support aerobic growth on purines as the sole carbon source (purine catabolism)
- Gene Name:
- xdhB
- Locus Tag:
- PA1523
- Molecular weight:
- 87.7 kDa
Reactions
| Xanthine + NAD(+) + H(2)O = urate + NADH. |
| Hypoxanthine + NAD(+) + H(2)O = xanthine + NADH. |
- General function:
- Not Available
- Specific function:
- Not Available
- Gene Name:
- paoA
- Locus Tag:
- PA1931
- Molecular weight:
- 18.3 kDa
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Specific, proton motive force-dependent high-affinity transporter for xanthine
- Gene Name:
- xanP
- Locus Tag:
- PA0352
- Molecular weight:
- 47.9 kDa