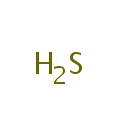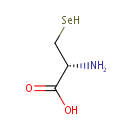
Search Results for compounds
Searching compounds for
returned 4373 results.
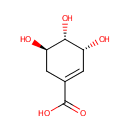
Shikimic acid (PAMDB000477)
IUPAC:
(3R,4S,5R)-3,4,5-trihydroxycyclohex-1-ene-1-carboxylic acid
CAS: 138-59-0
Description: Shikimic acid, more commonly known as its anionic form shikimate, is an important biochemical intermediate in plants and microorganisms. Its name comes from the Japanese flower shikimi (
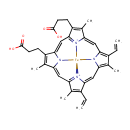
Heme (PAMDB000481)
IUPAC:
(1R)-15,19-bis(2-carboxyethyl)-5,10-diethenyl-4,9,14,20-tetramethyl-2???22,23???25-tetraaza-1-ferraoctacyclo[11.9.1.1?,??1?,??.0?,??0?????.0?????.0??,???pentacosa-2,4,6,8,10,12,14,16(23),17,19,21(24)-undecaene-2,23-bis(ylium)
CAS: 14875-96-8
Description: Heme B or haem B (also known as protoheme IX) is the most abundant heme in nature. Pseudomonas aeruginosa is known to produce 4 different hemes: protoheme IX (heme B), heme C, heme D, and siroheme. A heme or haem is a prosthetic group that consists of an iron atom contained in the center of a large heterocyclic organic ring called a porphyrin. Not all porphyrins contain iron, but a substantial fraction of porphyrin-containing metalloproteins have heme as their prosthetic subunit; these are known as hemoproteins. Generally, heme B is attached to the surrounding protein matrix (known as the apoprotein) through a single coordination bond between the heme iron and an amino-acid side-chain. When oxygen is bound the iron becomes hexacoordinated. Since the iron in heme B containing proteins is bound to the four nitrogens of the porphyrin (forming a plane) and a single electron donating atom of the protein, the iron is often in a pentacoordinate state.
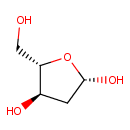
Deoxyribose (PAMDB000483)
IUPAC:
(2S,4R,5S)-5-(hydroxymethyl)oxolane-2,4-diol
CAS: 533-67-5
Description: Deoxyribose is an aldopentose, a monosaccharide containing five carbon atoms, and including an aldehyde functional group. It is derived from the pentose sugar ribose by the replacement of the hydroxyl group at the 2 position with hydrogen, leading to the net loss of an oxygen atom, and has chemical formula C5H10O4. In deoxyribose, the carbon furthest from the attached carbon is stripped of the oxygen atom in what would be a hydroxyl group in ribose. The common base adenine (a purine derivative) coupled to deoxyribose is called deoxyadenosine. The 5-triphosphate derivative of adenosine, commonly called adenosine triphosphate (ATP) is an important energy transport molecule in cells. (Wikipedia)
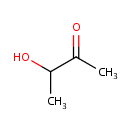
Acetoin (PAMDB000484)
IUPAC:
3-hydroxybutan-2-one
CAS: 513-86-0
Description: Acetoin is a chemical compound with the formula C4H8O2. It is a colourless or pale yellow to green yellow liquid with a pleasant buttery odour. It is used as a food flavoring and a fragrance. Acetoin is a product of fermentation. It is a component of the butanediol cycle in Pseudomonas aeruginosa. Pseudomonas aeruginosa pyruvate oxidase (POXEC) requires FAD both for the oxidative decarboxylation of pyruvate to acetate and CO2 and for the formation of acetoin from pyruvate and acetaldehyde. [PMID:8424670]
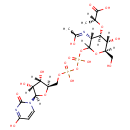
UDP-N-Acetylmuraminate (PAMDB000485)
IUPAC:
(2R)-2-{[(3R,4R,5S,6R)-2-({[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-5-hydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-4-yl]oxy}propanoic acid
CAS: Not Available
Description: UDP-N-acetylmuaminate is nucleoside diphosphate sugar which is formed from UDP-N-acetylglucosamine and phosphoenolpyruvate. It is a key component of peptidoglycan synthesis. The peptidoglycan synthesis pathway starts at the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-alpha-D-glucosamine, yielding the complete monomeric unit a lipid , also known as lipid . This final lipid intermediate is transferred through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks.
Hydrogen sulfide (PAMDB000486)
IUPAC:
hydrogen sulfide
CAS: 7783-06-4
Description: Hydrogen sulfide is a highly toxic and flammable gas. Because it is heavier than air it tends to accumulate at the bottom of poorly ventilated spaces. Although very pungent at first, it quickly deadens the sense of smell, so potential victims may be unaware of its presence until it is too late. H2S arises from virtually anywhere where elemental sulfur comes into contact with organic material, especially at high temperatures. Hydrogen sulfide is a covalent hydride chemically related to water (H2O) since oxygen and sulfur occur in the same periodic table group. It often results when bacteria break down organic matter in the absence of oxygen, such as in swamps, and sewers (alongside the process of anaerobic digestion). It also occurs in volcanic gases, natural gas and some well waters. It is also important to note that Hydrogen sulfide is a central participant in the sulfur cycle, the biogeochemical cycle of sulfur on earth. As mentioned above, sulfur-reducing and sulfate-reducing bacteria derive energy from oxidizing hydrogen or organic molecules in the absence of oxygen by reducing sulfur or sulfate to hydrogen sulfide. Other bacteria liberate hydrogen sulfide from sulfur-containing amino acids. Several groups of bacteria can use hydrogen sulfide as fuel, oxidizing it to elemental sulfur or to sulfate by using oxygen or nitrate as oxidant. The purple sulfur bacteria and the green sulfur bacteria use hydrogen sulfide as electron donor in photosynthesis, thereby producing elemental sulfur. (In fact, this mode of photosynthesis is older than the mode of cyanobacteria, algae and plants which uses water as electron donor and liberates oxygen).
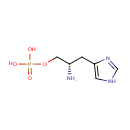
Histidinol phosphate (PAMDB000487)
IUPAC:
[(2S)-2-amino-3-(1H-imidazol-4-yl)propoxy]phosphonic acid
CAS: 25680-11-9
Description: Histidinol phosphate is a member of the chemical class known as Phosphoethanolamines. These are compounds containing a phosphate linked to the second carbon of an ethanolamine. L-histidinol phosphate (CHEBI:16996) has functional parent L-histidinol (CHEBI:16255) L-histidinol phosphate (CHEBI:16996) is a phosphoethanolamine (CHEBI:36711) L-histidinol phosphate (CHEBI:16996) is conjugate acid of L-histidinol phosphate (CHEBI:57980)
Selenocysteine (PAMDB000488)
IUPAC:
(2R)-2-amino-3-selanylpropanoic acid
CAS: 3614-08-2
Description: Selenocysteine is considered to be the 21st proteinogenic amino acid. It exists naturally in all kingdoms of life as a building block of selenoproteins. Selenocysteine is a cysteine analogue with a selenium-containing selenol group in place of the sulfur-containing thiol group. Selenocysteine is present in several enzymes (for example glutathione peroxidases, tetraiodothyronine 5' deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases,
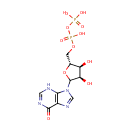
IDP (PAMDB000490)
IUPAC:
[({[(2R,3S,4R)-3,4-dihydroxy-5-(6-oxo-6,9-dihydro-3H-purin-9-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphonic acid
CAS: 86-04-4
Description: IDP is an inosine nucleotide containing a pyrophosphate group esterified to the C5 of the sugar moiety.
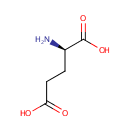
D-Glutamic acid (PAMDB000491)
IUPAC:
(2R)-2-aminopentanedioic acid
CAS: 6893-26-1
Description: D-glutamate is a key component of peptidoglycan synthesis. The peptidoglycan synthesis pathway starts at the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-alpha-D-glucosamine, yielding the complete monomeric unit a lipid , also known as lipid . This final lipid intermediate is transferred through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. There are two forms of glutamic acid found in nature: L-glutamic acid and D-glutamic acid. D-glutamic acid is found naturally primarily in the cell walls of certain bacteria - including Pseudomonas aeruginosa. D-glutamate is also present in certain foods e.g., soybeans and also arises from the turnover of the intestinal tract microflora, whose cell walls contain significant D-glutamate. Unlike other D-amino acids, D-glutamate is not oxidized by the D-amino acid oxidases, and therefore this detoxification pathway is not available for handling D-glutamate. Likewise, D-glutamic acid, when ingested, largely escapes most deamination reactions (unlike the L-counterpart). D-glutamate is the most potent natural inhibitor of glutathione synthesis identified to date.