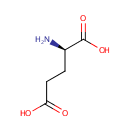
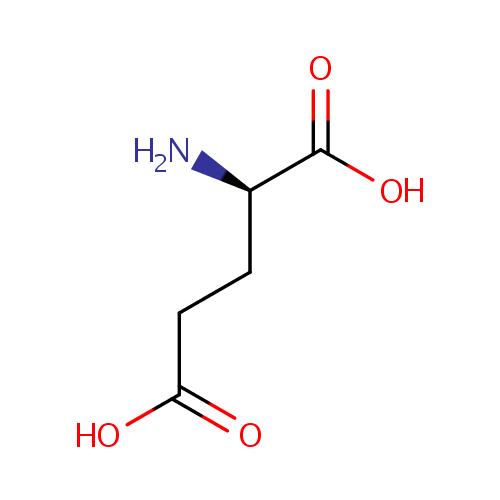
| References: |
- Ahlman B, Andersson K, Leijonmarck CE, Ljungqvist O, Hedenborg L, Wernerman J: Short-term starvation alters the free amino acid content of the human intestinal mucosa. Clin Sci (Lond). 1994 Jun;86(6):653-62. Pubmed: 7914846
- Bill JR, Mack DG, Falta MT, Maier LA, Sullivan AK, Joslin FG, Martin AK, Freed BM, Kotzin BL, Fontenot AP: Beryllium presentation to CD4+ T cells is dependent on a single amino acid residue of the MHC class II beta-chain. J Immunol. 2005 Nov 15;175(10):7029-37. Pubmed: 16272364
- Canal N, Nemni R: Autoimmunity and diabetic neuropathy. Clin Neurosci. 1997;4(6):371-3. Pubmed: 9358982
- Cuervo JH, Rodriguez B, Houghten RA: The Magainins: sequence factors relevant to increased antimicrobial activity and decreased hemolytic activity. Pept Res. 1988 Nov-Dec;1(2):81-6. Pubmed: 2980783
- Danke NA, Koelle DM, Yee C, Beheray S, Kwok WW: Autoreactive T cells in healthy individuals. J Immunol. 2004 May 15;172(10):5967-72. Pubmed: 15128778
- Emi M, Wilson DE, Iverius PH, Wu L, Hata A, Hegele R, Williams RR, Lalouel JM: Missense mutation (Gly----Glu188) of human lipoprotein lipase imparting functional deficiency. J Biol Chem. 1990 Apr 5;265(10):5910-6. Pubmed: 1969408
- Guariso G, Brotto F, Basso D, Alaggio R, Betterle C: Organ-specific autoantibodies in children with Helicobacter pylori infection. Helicobacter. 2004 Dec;9(6):622-8. Pubmed: 15610075
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Luo J, Kaplitt MG, Fitzsimons HL, Zuzga DS, Liu Y, Oshinsky ML, During MJ: Subthalamic GAD gene therapy in a Parkinson's disease rat model. Science. 2002 Oct 11;298(5592):425-9. Pubmed: 12376704
- Maechler P, Gjinovci A, Wollheim CB: Implication of glutamate in the kinetics of insulin secretion in rat and mouse perfused pancreas. Diabetes. 2002 Feb;51 Suppl 1:S99-102. Pubmed: 11815466
- Majid SM, Liss AS, You M, Bose HR: The suppression of SH3BGRL is important for v-Rel-mediated transformation. Oncogene. 2006 Feb 2;25(5):756-68. Pubmed: 16186799
- Mally MI, Cirulli V, Hayek A, Otonkoski T: ICA69 is expressed equally in the human endocrine and exocrine pancreas. Diabetologia. 1996 Apr;39(4):474-80. Pubmed: 8777998
- Meyer W, Poehling HM, Neurand K: Intraepidermal distribution of free amino acids in porcine skin. J Dermatol Sci. 1991 Sep;2(5):383-92. Pubmed: 1742249
- Nagai A, Suzuki Y, Baek SY, Lee KS, Lee MC, McLarnon JG, Kim SU: Generation and characterization of human hybrid neurons produced between embryonic CNS neurons and neuroblastoma cells. Neurobiol Dis. 2002 Oct;11(1):184-98. Pubmed: 12460557
- Nielsen C, Hansen D, Husby S, Jacobsen BB, Lillevang ST: No allelic variation in genes with high gliadin homology in patients with celiac disease and type 1 diabetes. Immunogenetics. 2004 Aug;56(5):375-8. Epub 2004 Aug 7. Pubmed: 15309343
- Persson H, Pelto-Huikko M, Metsis M, Soder O, Brene S, Skog S, Hokfelt T, Ritzen EM: Expression of the neurotransmitter-synthesizing enzyme glutamic acid decarboxylase in male germ cells. Mol Cell Biol. 1990 Sep;10(9):4701-11. Pubmed: 1697032
- Raj D, Langford M, Krueger S, Shelton M, Welbourne T: Regulatory responses to an oral D-glutamate load: formation of D-pyrrolidone carboxylic acid in humans. Am J Physiol Endocrinol Metab. 2001 Feb;280(2):E214-20. Pubmed: 11158923
- Roll U, Scheeser J, Standl E, Ziegler AG: Alterations of lymphocyte subsets in children of diabetic mothers. Diabetologia. 1994 Nov;37(11):1132-41. Pubmed: 7867885
- Wang M, Meng Z, Fu J: Synthesis and biodistribution of six novel 99mTc complexes of 2-hydroxybenzaldehyde-amino acid Schiff bases. Appl Radiat Isot. 2006 Feb;64(2):235-40. Pubmed: 16309915
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|