| Component |
|---|
| macromolecular complex |
| protein complex |
| proton-transporting ATP synthase complex, coupling factor F(o) |
| proton-transporting two-sector ATPase complex, proton-transporting domain |
| Function |
|---|
| cation transmembrane transporter activity |
| hydrogen ion transmembrane transporter activity |
| inorganic cation transmembrane transporter activity |
| ion transmembrane transporter activity |
| monovalent inorganic cation transmembrane transporter activity |
| substrate-specific transmembrane transporter activity |
| transmembrane transporter activity |
| transporter activity |
| Process |
|---|
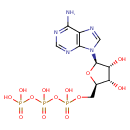
| ATP biosynthetic process |
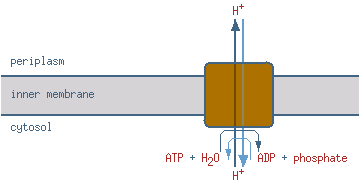
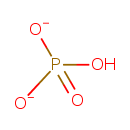
| ATP synthesis coupled proton transport |
| cellular nitrogen compound metabolic process |
| metabolic process |
| nitrogen compound metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| purine nucleoside triphosphate biosynthetic process |
| purine nucleotide biosynthetic process |
| purine nucleotide metabolic process |
| purine ribonucleoside triphosphate biosynthetic process |