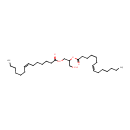| Identification |
| Name: |
Diacylglycerol kinase |
| Synonyms: |
- DAGK
- Diglyceride kinase
- DGK
|
| Gene Name: |
dgkA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in diacylglycerol kinase activity |
| Specific Function: |
Recycling of diacylglycerol produced during the turnover of membrane phospholipid |
| Cellular Location: |
Cell inner membrane; Multi-pass membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
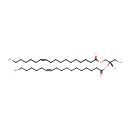
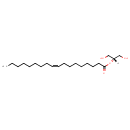
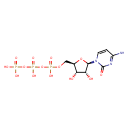
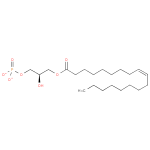
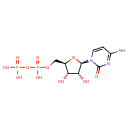
 | + | 1,2-Diacyl-sn-glycerol | + | 1,2-Diacyl-sn-glycerol | ↔ |  | + | 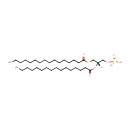 |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
| | | | | | | | | | | | |
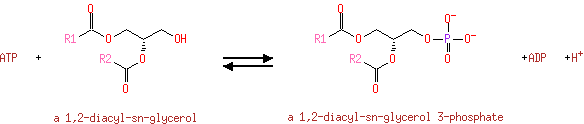
 | + | 1,2-diacylglycerol | → |  | + | 1,2-diacyl-sn-glycerol 3-phosphate |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB001512 | 1,2-Diacyl-sn-glycerol (didodecanoyl, n-C12:0) | MetaboCard | | PAMDB001513 | 1,2-Diacyl-sn-glycerol (dihexadec-9-enoyl, n-C16:1) | MetaboCard | | PAMDB001514 | 1,2-Diacyl-sn-glycerol (dihexadecanoyl, n-C16:0) | MetaboCard | | PAMDB001515 | 1,2-Diacyl-sn-glycerol (dioctadec-11-enoyl, n-C18:1) | MetaboCard | | PAMDB001517 | 1,2-Diacyl-sn-glycerol (ditetradec-7-enoyl, n-C14:1) | MetaboCard | | PAMDB001518 | 1,2-Diacyl-sn-glycerol (ditetradecanoyl, n-C14:0) | MetaboCard | | PAMDB001861 | 1,2-Dioctadecanoyl-sn-glycerol | MetaboCard | | PAMDB000148 | Adenosine triphosphate | MetaboCard | | PAMDB000345 | ADP | MetaboCard | | PAMDB003631 | DG(10:0/10:0/0:0) | MetaboCard | | PAMDB003632 | DG(10:0/12:0/0:0) | MetaboCard | | PAMDB003633 | DG(10:0/14:0/0:0) | MetaboCard | | PAMDB003634 | DG(10:0/15:0/0:0) | MetaboCard | | PAMDB003635 | DG(10:0/16:0/0:0) | MetaboCard | | PAMDB003636 | DG(10:0/16:1(9Z)/0:0) | MetaboCard | | PAMDB003637 | DG(10:0/18:0/0:0) | MetaboCard | | PAMDB003638 | DG(10:0/18:1(9Z)/0:0) | MetaboCard | | PAMDB003639 | DG(10:0/19:1(9Z)/0:0) | MetaboCard | | PAMDB003640 | DG(12:0/10:0/0:0) | MetaboCard | | PAMDB003641 | DG(12:0/12:0/0:0) | MetaboCard | | PAMDB003642 | DG(12:0/14:0/0:0) | MetaboCard | | PAMDB003643 | DG(12:0/15:0/0:0) | MetaboCard | | PAMDB003644 | DG(12:0/16:0/0:0) | MetaboCard | | PAMDB003645 | DG(12:0/16:1(9Z)/0:0) | MetaboCard | | PAMDB003646 | DG(12:0/18:0/0:0) | MetaboCard | | PAMDB003647 | DG(12:0/18:1(9Z)/0:0) | MetaboCard | | PAMDB003648 | DG(12:0/19:1(9Z)/0:0) | MetaboCard | | PAMDB003649 | DG(14:0/10:0/0:0) | MetaboCard | | PAMDB003650 | DG(14:0/12:0/0:0) | MetaboCard | | PAMDB003651 | DG(14:0/14:0/0:0) | MetaboCard | | PAMDB003652 | DG(14:0/15:0/0:0) | MetaboCard | | PAMDB003653 | DG(14:0/16:0/0:0) | MetaboCard | | PAMDB003654 | DG(14:0/16:1(9Z)/0:0) | MetaboCard | | PAMDB003655 | DG(14:0/18:0/0:0) | MetaboCard | | PAMDB003656 | DG(14:0/18:1(9Z)/0:0) | MetaboCard | | PAMDB003657 | DG(14:0/19:1(9Z)/0:0) | MetaboCard | | PAMDB003658 | DG(15:0/10:0/0:0) | MetaboCard | | PAMDB003659 | DG(15:0/12:0/0:0) | MetaboCard | | PAMDB003660 | DG(15:0/14:0/0:0) | MetaboCard | | PAMDB003661 | DG(15:0/15:0/0:0) | MetaboCard | | PAMDB003662 | DG(15:0/16:0/0:0) | MetaboCard | | PAMDB003663 | DG(15:0/16:1(9Z)/0:0) | MetaboCard | | PAMDB003664 | DG(15:0/18:0/0:0) | MetaboCard | | PAMDB003665 | DG(15:0/18:1(9Z)/0:0) | MetaboCard | | PAMDB003666 | DG(15:0/19:1(9Z)/0:0) | MetaboCard | | PAMDB003667 | DG(16:0/10:0/0:0) | MetaboCard | | PAMDB003668 | DG(16:0/12:0/0:0) | MetaboCard | | PAMDB003669 | DG(16:0/14:0/0:0) | MetaboCard | | PAMDB003670 | DG(16:0/15:0/0:0) | MetaboCard | | PAMDB003671 | DG(16:0/16:0/0:0) | MetaboCard | | PAMDB003672 | DG(16:0/16:1(9Z)/0:0) | MetaboCard | | PAMDB003673 | DG(16:0/18:0/0:0) | MetaboCard | | PAMDB003674 | DG(16:0/18:1(9Z)/0:0) | MetaboCard | | PAMDB003675 | DG(16:0/19:1(9Z)/0:0) | MetaboCard | | PAMDB003676 | DG(16:1(9Z)/10:0/0:0) | MetaboCard | | PAMDB003677 | DG(16:1(9Z)/12:0/0:0) | MetaboCard | | PAMDB003678 | DG(16:1(9Z)/14:0/0:0) | MetaboCard | | PAMDB003679 | DG(16:1(9Z)/15:0/0:0) | MetaboCard | | PAMDB003680 | DG(16:1(9Z)/16:0/0:0) | MetaboCard | | PAMDB003681 | DG(16:1(9Z)/16:1(9Z)/0:0) | MetaboCard | | PAMDB003682 | DG(16:1(9Z)/18:0/0:0) | MetaboCard | | PAMDB003683 | DG(16:1(9Z)/18:1(9Z)/0:0) | MetaboCard | | PAMDB003684 | DG(16:1(9Z)/19:1(9Z)/0:0) | MetaboCard | | PAMDB003685 | DG(18:0/10:0/0:0) | MetaboCard | | DG(18:0/12:0/0:0) | MetaboCard | | PAMDB003687 | DG(18:0/14:0/0:0) | MetaboCard | | PAMDB003688 | DG(18:0/15:0/0:0) | MetaboCard | | PAMDB003689 | DG(18:0/16:0/0:0) | MetaboCard | | PAMDB003690 | DG(18:0/16:1(9Z)/0:0) | MetaboCard | | PAMDB003691 | DG(18:0/18:0/0:0) | MetaboCard | | PAMDB003692 | DG(18:0/18:1(9Z)/0:0) | MetaboCard | | PAMDB003693 | DG(18:0/19:1(9Z)/0:0) | MetaboCard | | PAMDB003694 | DG(18:1(9Z)/10:0/0:0) | MetaboCard | | PAMDB003695 | DG(18:1(9Z)/12:0/0:0) | MetaboCard | | PAMDB003696 | DG(18:1(9Z)/14:0/0:0) | MetaboCard | | PAMDB003697 | DG(18:1(9Z)/15:0/0:0) | MetaboCard | | PAMDB003698 | DG(18:1(9Z)/16:0/0:0) | MetaboCard | | PAMDB003699 | DG(18:1(9Z)/16:1(9Z)/0:0) | MetaboCard | | PAMDB003700 | DG(18:1(9Z)/18:0/0:0) | MetaboCard | | PAMDB003701 | DG(18:1(9Z)/18:1(9Z)/0:0) | MetaboCard | | PAMDB003702 | DG(18:1(9Z)/19:1(9Z)/0:0) | MetaboCard | | PAMDB003703 | DG(19:1(9Z)/10:0/0:0) | MetaboCard | | PAMDB003704 | DG(19:1(9Z)/12:0/0:0) | MetaboCard | | PAMDB003705 | DG(19:1(9Z)/14:0/0:0) | MetaboCard | | PAMDB003706 | DG(19:1(9Z)/15:0/0:0) | MetaboCard | | PAMDB003707 | DG(19:1(9Z)/16:0/0:0) | MetaboCard | | PAMDB003708 | DG(19:1(9Z)/16:1(9Z)/0:0) | MetaboCard | | PAMDB003709 | DG(19:1(9Z)/18:0/0:0) | MetaboCard | | PAMDB003710 | DG(19:1(9Z)/18:1(9Z)/0:0) | MetaboCard | | PAMDB003711 | DG(19:1(9Z)/19:1(9Z)/0:0) | MetaboCard | | PAMDB001633 | Hydrogen ion | MetaboCard | | PAMDB000169 | PA(16:0/16:0) | MetaboCard |
|
| GO Classification: |
| Component |
|---|
| cell part | | membrane | | Function |
|---|
| catalytic activity | | diacylglycerol kinase activity | | kinase activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| metabolic process | | organophosphate metabolic process | | phospholipid biosynthetic process | | phospholipid metabolic process |
|
| Gene Properties |
| Locus tag: |
PA3603 |
| Strand: |
- |
| Entrez Gene ID: |
880149 |
| Accession: |
NP_252293.1 |
| GI: |
15598799 |
| Sequence start: |
4037945 |
| Sequence End: |
4038316 |
| Sequence Length: |
371 |
| Gene Sequence: |
>PA3603
GTGTCGCCTTCCCCCTTCAAAGGCCAGACCGGCCTCAAGCGTATCCTCAACGCCACCGGCTATTCCCTCGCAGGTTTCCTCGCCGCCTTCCGCGGTGAAGCCGCCTTCCGCCAACTGGTGCTGCTCAACGTGGTGCTGATCCCGGTGGCCTTCCTCCTCGACGTCAGTCGCGGCGAACGCGCGCTGATGATCGCGGTGTGCCTGCTGGCGCTGATCGTCGAGCTGCTCAACTCGGCGATCGAGGCCACCGTCGACCGCGTCTCGCTGGAACGCCACCCTCTGTCGAAGAATGCCAAGGACATGGGCAGCGCCGCGCAGTTCGTGGCCCTCACCGTGATCACCGTGACCTGGGCGACCATCCTGCTGGGCTGA |
| Protein Properties |
| Protein Residues: |
123 |
| Protein Molecular Weight: |
13.1 kDa |
| Protein Theoretical pI: |
10.03 |
| Hydropathicity (GRAVY score): |
0.762 |
| Charge at pH 7 (predicted): |
3.19 |
| Protein Sequence: |
>PA3603
MSPSPFKGQTGLKRILNATGYSLAGFLAAFRGEAAFRQLVLLNVVLIPVAFLLDVSRGERALMIAVCLLALIVELLNSAIEATVDRVSLERHPLSKNAKDMGSAAQFVALTVITVTWATILLG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3603 and its homologs
|