| Identification |
| Name: |
Tetraacyldisaccharide 4'-kinase |
| Synonyms: |
|
| Gene Name: |
lpxK |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in ATP binding |
| Specific Function: |
Transfers the gamma-phosphate of ATP to the 4'-position of a tetraacyldisaccharide 1-phosphate intermediate (termed DS-1- P) to form tetraacyldisaccharide 1,4'-bis-phosphate (lipid IVA) |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
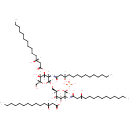
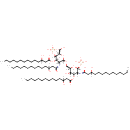
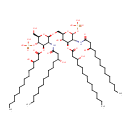
(2-N,3-O-bis(3-hydroxytetradecanoyl)-beta-D-glucosaminyl)-(1->6)-(2-N,3-O-bis(3-hydroxytetradecanoyl)-beta-D-glucosaminyl phosphate) | + |  | → | Adenosine diphosphate | + |  | + | 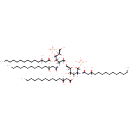 | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB003481 | (2-N,3-O-bis(3-Hydroxytetradecanoyl)-4-O-phosphono-beta-D-glucosaminyl)-(1->6)-(2-N,3-O-bis(3-hydroxytetradecanoyl)-beta-D-glucosaminyl phosphate) | MetaboCard | | PAMDB000905 | 2,3,2',3'-Tetrakis(3-hydroxytetradecanoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1-phosphate | MetaboCard | | PAMDB000906 | 2,3,2'3'-Tetrakis(3-hydroxytetradecanoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate | MetaboCard | | PAMDB001545 | 2,3,2'3'-Tetrakis(beta-hydroxymyristoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate | MetaboCard | | PAMDB000148 | Adenosine triphosphate | MetaboCard | | PAMDB000345 | ADP | MetaboCard | | PAMDB001633 | Hydrogen ion | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | catalytic activity | | kinase activity | | nucleoside binding | | purine nucleoside binding | | tetraacyldisaccharide 4'-kinase activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| lipid A biosynthetic process | | metabolic process | | organophosphate metabolic process | | phospholipid biosynthetic process | | phospholipid metabolic process |
|
| Gene Properties |
| Locus tag: |
PA2981 |
| Strand: |
- |
| Entrez Gene ID: |
880503 |
| Accession: |
NP_251671.1 |
| GI: |
15598177 |
| Sequence start: |
3338679 |
| Sequence End: |
3339677 |
| Sequence Length: |
998 |
| Gene Sequence: |
>PA2981
ATGTCGTTCTCCGAGCGACTGCTCGCCGCCTGGTACCAGGGGCATCCGGCGCTGGCGCTGCTGCGTCCGCTGGAGGCTCTCTATCGTCGGGTGGCGAACGGCCGCCGGGCGGACTTCCTGTCCGGGCGCAAGCCCGCCTACCGGGCACCGCTGCCGGTGCTGGTGGTAGGCAACATCACCGTCGGCGGAACCGGCAAGACGCCGATGATCCTCTGGATGATCGAGCACTGCAGGGCTCGCGGCTTGCGGGTTGGCGTGATCAGCCGCGGCTATGGTGCCCGGCCGCCCACCACGCCGTGGCGGGTGCGGGCCGAGCAGGACGCCGCGGAGGCCGGCGACGAACCGCTGATGATCGTCCGGCGCAGCGGCGTGCCGCTGATGATCGACCCGGATCGTCCGCGCGCCCTGCAGGCGCTGCTCGCCGAAGAGCAGTTGGACCTGGTCCTCTGCGACGATGGCTTGCAGCACTATCGCCTGGCACGCGATCTGGAGCTGGTGCTGATCGATGCCGCGCGCGGTCTCGGCAATGGTCGTTGCCTGCCGGCCGGTCCGTTGCGCGAACCGGCGGAGCGTCTGGAAAGCGTCGACGCGCTGCTTTACAACGGCGCCGACGAGGATCCTGACGGCGGTTACGCATTCCGCTTGCAGCCTACTGCGCTGATCAACCTGCAGAGCGGCGAGCGCCGACCGCTGGAGCATTTTCCCGCGGGCCAGGAGGTCCATGCCCTGGCCGGGATCGGCAATCCGCAGCGTTTCTTCAGGACGCTCGAGGCGCTACACTGGCGGGCGATTCCGCATGCCTTCCCGGATCACGCGACCTACACGGCGGCCGAACTGGCGTTCAGCCCGCCGCTGCCGCTGCTGATGACCGAGAAGGATGCCGTTAAATGCCGGGCTTTCGCAGCGGCCGACTGGTGGTACCTGGCGGTCGATGCGGTGCCTTCCCCAGCATTCGTCGCCTGGTTCGACGCCCGACTCGAGCATCTCCTGGCGCGCTGA |
| Protein Properties |
| Protein Residues: |
332 |
| Protein Molecular Weight: |
36.7 kDa |
| Protein Theoretical pI: |
7.29 |
| Hydropathicity (GRAVY score): |
-0.119 |
| Charge at pH 7 (predicted): |
1.05 |
| Protein Sequence: |
>PA2981
MSFSERLLAAWYQGHPALALLRPLEALYRRVANGRRADFLSGRKPAYRAPLPVLVVGNITVGGTGKTPMILWMIEHCRARGLRVGVISRGYGARPPTTPWRVRAEQDAAEAGDEPLMIVRRSGVPLMIDPDRPRALQALLAEEQLDLVLCDDGLQHYRLARDLELVLIDAARGLGNGRCLPAGPLREPAERLESVDALLYNGADEDPDGGYAFRLQPTALINLQSGERRPLEHFPAGQEVHALAGIGNPQRFFRTLEALHWRAIPHAFPDHATYTAAELAFSPPLPLLMTEKDAVKCRAFAAADWWYLAVDAVPSPAFVAWFDARLEHLLAR |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA2981 and its homologs
|