| Identification |
| Name: |
Periplasmic beta-glucosidase |
| Synonyms: |
- Beta-D-glucoside glucohydrolase
- Cellobiase
- Gentiobiase
|
| Gene Name: |
bglX |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in hydrolase activity, hydrolyzing O-glycosyl compounds |
| Specific Function: |
Hydrolysis of terminal, non-reducing beta-D- glucosyl residues with release of beta-D-glucose |
| Cellular Location: |
Periplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
| | |
Cellulose | + |  | ↔ | Cellulose | + |  |
| | |
D-Glucoside | + |  | ↔ | ROH | + | 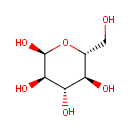 |
| | |
Cyanoglycoside | + |  | ↔ | Cyanohydrin | + | 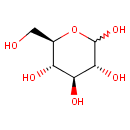 |
| | |
cis-beta-D-Glucosyl-2-hydroxycinnamate | + |  | ↔ | cis-2-Hydroxycinnamate | + |  |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
a β-D glucoside | + |  | ? | a non glucosylated glucose acceptor | + |  |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | hydrolase activity | | hydrolase activity, acting on glycosyl bonds | | hydrolase activity, hydrolyzing O-glycosyl compounds | | Process |
|---|
| carbohydrate metabolic process | | metabolic process | | primary metabolic process |
|
| Gene Properties |
| Locus tag: |
PA1726 |
| Strand: |
- |
| Entrez Gene ID: |
877660 |
| Accession: |
NP_250417.1 |
| GI: |
15596923 |
| Sequence start: |
1866241 |
| Sequence End: |
1868535 |
| Sequence Length: |
2294 |
| Gene Sequence: |
>PA1726
ATGAACAGGCTGTGTGTACTGGGGCTTCTCATCGGGCTGAGCGGGAGCCCGGCACTGGCGGCTGGCCAGGGCGAATCCGGCAATCCGCCGGCCACCGACAAGGAGCGTTTCATCGCCAGCCTGATGGCTCGCATGAGCAACGCGGAAAAGATCGGCCAGCTACGCCTGGTCAGCGTCGGCGCCGACCATCCCAAGGAAGCGCTGATGGCGGATATCCGCGCCGGCAAGGTCGGCGCCATATTCAATACCGTGACCCGCCCGGACATTCGCGCCATGCAGGACCAGGTCCGCCATAGCCGCCTGAAGATCCCGCTGTTCCATGCCTACGACGTGGCCCACGGGCACCGCACCATCTTTCCCATCAGCCTCGGCCTGGCCGCCAGCTGGGACCCCGAGGTGGTCGCCCGCAGCGCGCGGATCTCCGCCCTCGAAGCCAGCGCCGACGGGCTCGACATGAGCTTCTCGCCTATGGTCGACATCACCCGCGACGCGCGCTGGGGGCGGGTCTCGGAAGGCTTCGGCGAAGACACCTACCTGACCTCGCTGCTGTCCGGCGTGATGGTCCGTGCCTACCAGGGCTCCAACCTGGCCGCCCCGGACAGCATCATGGCGGCGGTCAAGCACTTCGCCCTGTACGGCGCGGCCGAAGGCGGCCGCGACTACAACACCGTGGACATGAGCCTGCCGCGCATGTTCCAGGACTACCTGCCGCCCTACAAGGCCGCGGTGGACGCCGGCGCCGGCGCGGTGATGGTCTCGCTGAACACCATCAACGGCGTGCCCGCCACCGCCAACCGCTGGCTGCTGACCGACCTGTTGCGCCAGCAGTGGGGCTTCAAGGGGCTGACCATCAGTGACCACGGCGCGGTGAAGGAACTGATCAAGCACGGCCTCGCCGGCAACGAGCGGGACGCCACGCGCCTGGCGATCCAGGCCGGTGTCGACATGAACATGAACGACGATCTCTACTCGACCTGGCTACCGAAGCTGCTGGCGGCCGGCGAGATCGACCAGGCCGACATCGATCGCGCCTGCCGCGACGTGCTGGCGGCGAAGTACGACCTCGGCCTGTTCGCCGATCCCTACCGGCGCCTGGGCAAGCCGGACGACCCGCCGTTCGACACCAACGCCGAGAGCCGCCTGCACCGCCAGGCCGCCCGCGAGGTGGCTCGCGAAGGACTGGTCCTGCTGAAGAACCGCGACGGTCTGCTGCCGCTGAAGAAACAGGGGCGGATCGCCGTGATCGGCCCCCTGGCCAAGAGCCAGCGCGACGTCATCGGCAGTTGGTCGGCAGCCGGCGTGCCACGCCAGGCGGTAACCGTCTACCAGGGCCTGGCCAATGCCGTCGGCGAGCGTGCCACCCTGCTCTACGCCAAGGGTGCCAACGTCAGTGGCGACCAGGCGATCCTCGACTACCTGAACAGCTACAACCCGGAAGTCGAGGTCGATCCGCGCTCCGCCGAGGCGATGCTCGAGGAGGCCTTGCGCACCGCGCGCGACGCCGACCTGGTGGTCGCCGTGGTCGGCGAATCGCAGGGCATGGCCCACGAGGCCTCCAGTCGCACCGACCTGCGCATCCCGGCCAGCCAGCGGCGGCTGCTGAAGGCACTGAAGGCCACCGGCAAGCCGCTGGTGCTGGTGCTGATGAACGGCCGACCGCTGTCGCTGGGCTGGGAGCAGGAAAACGCCGACGCGATCCTGGAAACCTGGTTCAGTGGCACCGAAGGCGGCAACGCCATCGCCGACGTGCTGTTCGGCGAGCACAACCCCAGCGGCAAGCTGACCATGTCCTTCCCGCGCTCGGTGGGCCAGGTTCCGGTTTACTACAACCACCTGAACACCGGCCGGCCGATGGACCACGACAACCCCGGCAAATACACCTCGCGCTATTTCGACGAAGCCAACGGCCCGCTCTACCCGTTCGGCTACGGCCTCAGCTACACCGAGTTCAGTCTCTCGCCCCTGCGCCTGTCCAGCGAACGGCTGGCCCGCGGCGCTACCCTGGAAGCGCGGGTCACGCTCAGTAACAGCGGCAAGCGCGCCGGCGCCACGGTGGTCCAGCTTTATCTGCAGGACCCGGTCGCCAGCCTCAGCCGTCCGGTGAAGGAACTGCGCGGGTTCCGCAAGGTGATGCTGGAGCCCGGCGAATCGCGGGAGATCGTCTTCCGCCTCGGCGAGGCAGACCTAAAGTTCTACGACAGCCAGCTCAGGCACACGGCCGAGCCGGGCGAGTTCAAGGTCTTCGTCGGCCTGGATTCGGCGCAAACCGAAAGCCGCAGCTTCACCCTGCTCTGA |
| Protein Properties |
| Protein Residues: |
764 |
| Protein Molecular Weight: |
83 kDa |
| Protein Theoretical pI: |
7.06 |
| Hydropathicity (GRAVY score): |
-0.206 |
| Charge at pH 7 (predicted): |
0.34 |
| Protein Sequence: |
>PA1726
MNRLCVLGLLIGLSGSPALAAGQGESGNPPATDKERFIASLMARMSNAEKIGQLRLVSVGADHPKEALMADIRAGKVGAIFNTVTRPDIRAMQDQVRHSRLKIPLFHAYDVAHGHRTIFPISLGLAASWDPEVVARSARISALEASADGLDMSFSPMVDITRDARWGRVSEGFGEDTYLTSLLSGVMVRAYQGSNLAAPDSIMAAVKHFALYGAAEGGRDYNTVDMSLPRMFQDYLPPYKAAVDAGAGAVMVSLNTINGVPATANRWLLTDLLRQQWGFKGLTISDHGAVKELIKHGLAGNERDATRLAIQAGVDMNMNDDLYSTWLPKLLAAGEIDQADIDRACRDVLAAKYDLGLFADPYRRLGKPDDPPFDTNAESRLHRQAAREVAREGLVLLKNRDGLLPLKKQGRIAVIGPLAKSQRDVIGSWSAAGVPRQAVTVYQGLANAVGERATLLYAKGANVSGDQAILDYLNSYNPEVEVDPRSAEAMLEEALRTARDADLVVAVVGESQGMAHEASSRTDLRIPASQRRLLKALKATGKPLVLVLMNGRPLSLGWEQENADAILETWFSGTEGGNAIADVLFGEHNPSGKLTMSFPRSVGQVPVYYNHLNTGRPMDHDNPGKYTSRYFDEANGPLYPFGYGLSYTEFSLSPLRLSSERLARGATLEARVTLSNSGKRAGATVVQLYLQDPVASLSRPVKELRGFRKVMLEPGESREIVFRLGEADLKFYDSQLRHTAEPGEFKVFVGLDSAQTESRSFTLL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA1726 and its homologs
|