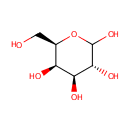
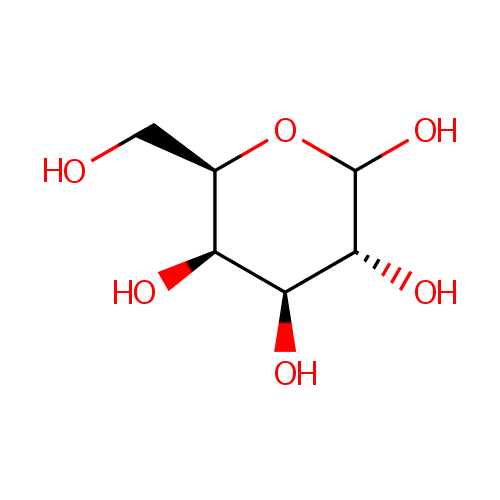
D-Galactose (PAMDB000054)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000054 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | D-Galactose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | D-Galactose is an aldohexose that occurs naturally in the D-form in lactose, cerebrosides, gangliosides, and mucoproteins. D-Galactose is an energy-providing nutrient and also a necessary basic substrate for the biosynthesis of many macromolecules. Metabolic pathways for D-Galactose are important not only for the provision of these pathways but also for the prevention of D-Galactose and D-Galactose metabolite accumulation. The main source of D-Galactose is lactose in the milk of mammals, but it can also be found in some fruits and vegetables. Utilization of D-Galactose in all living cells is initiated by the phosphorylation of the hexose by the enzyme galactokinase (E.C. 2.7.1.6) (GALK) to form D-Galactose-1-phosphate. In the presence of D-Galactose-1-phosphate uridyltransferase (E.C. 2.7.7.12) (GALT) D-Galactose-1-phosphate is exchanged with glucose-1-phosphate in UDP-glucose to form UDP-galactose. Glucose-1-phosphate will then enter the glycolytic pathway for energy production. (PMID: 15256214, 11020650, 10408771) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H12O6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 180.1559 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 180.063388116 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | WQZGKKKJIJFFOK-SVZMEOIVSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h2-11H,1H2/t2-,3+,4+,5-,6?/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 59-23-4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3R,4S,5R,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (+)-galactose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monosaccharides. These are compounds containing one carbohydrate unit not glycosidically linked to another such unit, and no set of two or more glycosidically linked carbohydrate units. Monosaccharides have the general formula CnH2nOn. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 170 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + D-Galactose > ADP + D-Galactose + Hydrogen ion + Phosphate Adenosine triphosphate + Water + D-Galactose > ADP + D-Galactose + Hydrogen ion + Phosphate Adenosine triphosphate + D-Galactose + Alpha-D-Galactose <> ADP + Galactose 1-phosphate + Hydrogen ion Water + alpha-Lactose > D-Galactose + D-Glucose beta-D-Galactose > D-Galactose Galactose 1-phosphate + Water > D-Galactose + Phosphate Water + Melibiose > D-Galactose + D-Glucose Adenosine triphosphate + D-Galactose <> ADP + Galactose 1-phosphate Galactan + Water <> D-Galactose + Galactan alpha-Lactose + Water <> alpha-D-Glucose + D-Galactose beta-D-Galactosyl-1,4-beta-D-glucosylceramide + Water <> Glucosylceramide + D-Galactose Digalactosylceramide + Water <> Galactosylceramide + D-Galactose Digalactosyl-diacylglycerol + Water <> 1,2-Diacyl-3-beta-D-galactosyl-sn-glycerol + D-Galactose D-Gal alpha 1->6D-Gal alpha 1->6D-Glucose + Water <> D-Galactose + Melibiose Water + Globotriaosylceramide <> D-Galactose + Lactosylceramide Melibiose + Water <> D-Galactose + D-Glucose Lactose + Water <> D-Glucose + D-Galactose D-Galactose + NAD 3-keto-β-D-galactose + NADH + Hydrogen ion More...-->-->D-Galactose <> D-Galactose D-Galactose <> D-Galactose Water + Melibiose > D-Galactose + b-D-Glucose Water + alpha-Lactose > D-Galactose + b-D-Glucose D-Galactose + Adenosine triphosphate > Hydrogen ion + Galactose 1-phosphate + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Avigad, Gad. Synthesis of D-galactose-6-t and D-galactosides-6-t. Carbohydrate Research (1967), 3(4), 430-4. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in hydrolase activity, hydrolyzing O-glycosyl compounds
- Specific function:
- Hydrolysis of terminal, non-reducing beta-D- glucosyl residues with release of beta-D-glucose
- Gene Name:
- bglX
- Locus Tag:
- PA1726
- Molecular weight:
- 83 kDa
Reactions
| Hydrolysis of terminal, non-reducing beta-D-glucosyl residues with release of beta-D-glucose. |