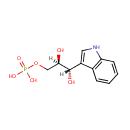| Identification |
| Name: |
Tryptophan synthase beta chain |
| Synonyms: |
Not Available |
| Gene Name: |
trpB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in catalytic activity |
| Specific Function: |
The beta subunit is responsible for the synthesis of L- tryptophan from indole and L-serine |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
- Glycine, serine and threonine metabolism pae00260
- Metabolic pathways pae01100
- Phenylalanine, tyrosine and tryptophan biosynthesis pae00400
- Tryptophan metabolism pae00380
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
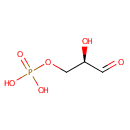
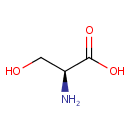 | + | 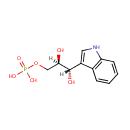 | → | 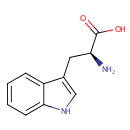 | + | glyceraldehyde 3-phosphate | + |  |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| binding | | carbon-oxygen lyase activity | | catalytic activity | | cofactor binding | | hydro-lyase activity | | lyase activity | | pyridoxal phosphate binding | | tryptophan synthase activity | | Process |
|---|
| cellular amino acid and derivative metabolic process | | cellular amino acid derivative metabolic process | | cellular biogenic amine metabolic process | | cellular metabolic process | | indolalkylamine metabolic process | | metabolic process | | tryptophan metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0036 |
| Strand: |
- |
| Entrez Gene ID: |
879212 |
| Accession: |
NP_248726.1 |
| GI: |
15595234 |
| Sequence start: |
37890 |
| Sequence End: |
39098 |
| Sequence Length: |
1208 |
| Gene Sequence: |
>PA0036
ATGACTTCCTATCGCAACGGCCCCGACGCCAAGGGCCTGTTCGGCCGCTTCGGCGGCCAGTACGTCGCCGAGACCCTGATGCCGCTGATCCTCGACCTCGCCCGCGAGTACGAGAAGGCCAAGGACGACCCGGCGTTCCAGGAGGAACTGGCCTACTTCCAGCGCGACTACGTCGGTCGACCGAGCCCGCTGTACTTCGCCGAGCGCCTGACCGAGCACTGCGGCGGGGCGAAGATCTACCTCAAGCGCGAGGAGCTGAACCATACCGGCGCGCACAAGATCAACAACTGCATCGGCCAGATCCTCCTGGCCCGGCGCATGGGCAAGAAACGCATCATCGCCGAGACCGGCGCCGGCATGCACGGCGTGGCCACTGCCACCGTCGCCGCGCGCTTCGGCCTGCAGTGCGTGATCTACATGGGCACCACCGACATCGACCGGCAGCAGGCCAACGTCTTCCGCATGAAGCTGCTGGGCGCCGAGGTGATCCCGGTGACCGCCGGCACCGGTACCCTGAAGGACGCCATGAACGAGGCGCTGCGCGACTGGGTGACCAACGTCGACAGCACCTTCTACCTGATCGGCACGGTCGCCGGCCCGCATCCGTACCCGGCGATGGTCCGCGACTTCCAGGCGGTGATCGGCAAGGAAACCCGCGAGCAACTGGCCGAGAAGGAAGGGCGCCTGCCCGATTCGCTGGTCGCCTGCATCGGCGGCGGCTCCAACGCCATGGGCCTGTTCCACCCGTTCCTCGACGACGCCGGGGTGCAGATCGTCGGCGTGGAAGCCGCCGGCCACGGCATCGACACCGGCAAGCACGCGGCCAGCCTGAACGGCGGGGTTCCCGGCGTGCTGCACGGCAACCGCACCTTCCTGCTGCAGGACGCGGATGGCCAGATCATCGACGCACACTCCATCTCCGCCGGCCTCGACTATCCCGGCATCGGCCCGGAACACGCCTGGCTGCACGACACCGGCCGCGTCGAGTACACCTCGATCACCGACGACGAAGCCCTGGAGGCCTTCCACACCTGCTGCCGCCTCGAAGGCATCATCCCGGCACTGGAAAGCTCCCATGCCCTGGCCGAGGTCTTCAAGCGTGCGCCCAGCCTGCCCAAGGAGCACATCATGGTGGTGAACCTGTCCGGTCGCGGCGACAAGGACATGCAGACCGTCATGCACCACATGCAACAGGAGTCGAAAGCATGA |
| Protein Properties |
| Protein Residues: |
402 |
| Protein Molecular Weight: |
43.7 kDa |
| Protein Theoretical pI: |
6.26 |
| Hydropathicity (GRAVY score): |
-0.203 |
| Charge at pH 7 (predicted): |
-7.08 |
| Protein Sequence: |
>PA0036
MTSYRNGPDAKGLFGRFGGQYVAETLMPLILDLAREYEKAKDDPAFQEELAYFQRDYVGRPSPLYFAERLTEHCGGAKIYLKREELNHTGAHKINNCIGQILLARRMGKKRIIAETGAGMHGVATATVAARFGLQCVIYMGTTDIDRQQANVFRMKLLGAEVIPVTAGTGTLKDAMNEALRDWVTNVDSTFYLIGTVAGPHPYPAMVRDFQAVIGKETREQLAEKEGRLPDSLVACIGGGSNAMGLFHPFLDDAGVQIVGVEAAGHGIDTGKHAASLNGGVPGVLHGNRTFLLQDADGQIIDAHSISAGLDYPGIGPEHAWLHDTGRVEYTSITDDEALEAFHTCCRLEGIIPALESSHALAEVFKRAPSLPKEHIMVVNLSGRGDKDMQTVMHHMQQESKA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0036 and its homologs
|